Abstract
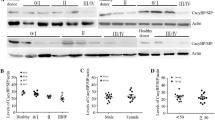
Hairy and enhancer of split homolog-1 (HES1), regulated by the Notch, has been reported to play important roles in the immune response and cancers, such as leukemia. In this study, we aim to explore the effect of HES1-mediated Notch1 signaling pathway in chronic lymphocytic leukemia (CLL). Reverse transcription quantitative polymerase chain reaction and Western blot assay were conducted to determine the expression of HES1, Notch1, and PTEN in B lymphocytes of peripheral blood samples of 60 CLL patients. We used lentivirus-mediated overexpression or silencing of HES1 and the Notch1 signaling pathway inhibitor, MW167, to detect the interaction among HES1, Notch1, and PTEN in CLL MEC1 and HG3 cells. MTT assay and flow cytometry were employed for detection of biological behaviors of CLL cells. HES1 and Notch1 showed high expression, but PTEN displayed low expression in B lymphocytes of peripheral blood samples of patients with CLL in association with poor prognosis. HES1 bound to the promoter region of PTEN and reduced PTEN expression. Overexpression of HES1 activated the Notch1 signaling pathway, thus promoting the proliferation of CLL cells, increasing the proportion of cells arrested at the S phase and limiting the apoptosis of CLL cells. Collectively, HES1 can promote activation of the Notch1 signaling pathway to cause PTEN transcription inhibition and the subsequent expression reduction, thereby promoting the proliferation and inhibiting the apoptosis of CLL cells.








Similar content being viewed by others
References
Savvopoulos, S., Misener, R., Panoskaltsis, N., Pistikopoulos, E. N., & Mantalaris, A. (2016). A personalized framework for dynamic modeling of disease trajectories in chronic lymphocytic leukemia. IEEE Transactions on Biomedical Engineering, 63, 2396–2404.
Wiestner, A. (2012). Emerging role of kinase-targeted strategies in chronic lymphocytic leukemia. Blood, 120, 4684–4691.
Hus, I., Salomon-Perzynski, A., & Robak, T. (2020). The up-to-date role of biologics for the treatment of chronic lymphocytic leukemia. Expert Opinion on Biological Therapy, 20, 799–812.
Stauder, R., Eichhorst, B., Hamaker, M. E., Kaplanov, K., Morrison, V. A., Osterborg, A., Poddubnaya, I., Woyach, J. A., Shanafelt, T., Smolej, L., Ysebaert, L., & Goede, V. (2017). Management of chronic lymphocytic leukemia (CLL) in the elderly: A position paper from an international Society of Geriatric Oncology (SIOG) Task Force. Annals of Oncology, 28, 218–227.
Eichhorst, B., Cramer, P., & Hallek, M. (2016). Initial therapy of chronic lymphocytic leukemia. Seminars in Oncology, 43, 241–250.
Lee, J. C., & Lamanna, N. (2020). Is there a role for chemotherapy in the era of targeted therapies? Current Hematologic Malignancy Reports, 15, 72–82.
Wu, Y., Gong, L., Xu, J., Mou, Y., Xu, X., & Qian, Z. (2017). The clinicopathological significance of HES1 promoter hypomethylation in patients with colorectal cancer. Oncotargets and Therapy, 10, 5827–5834.
Nakahara, F., Sakata-Yanagimoto, M., Komeno, Y., Kato, N., Uchida, T., Haraguchi, K., Kumano, K., Harada, Y., Harada, H., Kitaura, J., Ogawa, S., Kurokawa, M., Kitamura, T., & Chiba, S. (2010). Hes1 immortalizes committed progenitors and plays a role in blast crisis transition in chronic myelogenous leukemia. Blood, 115, 2872–2881.
Monahan, P., Rybak, S., & Raetzman, L. T. (2009). The notch target gene HES1 regulates cell cycle inhibitor expression in the developing pituitary. Endocrinology, 150, 4386–4394.
Ferrante, F., Giaimo, B. D., Bartkuhn, M., Zimmermann, T., Close, V., Mertens, D., Nist, A., Stiewe, T., Meier-Soelch, J., Kracht, M., Just, S., Kloble, P., Oswald, F., & Borggrefe, T. (2020). HDAC3 functions as a positive regulator in Notch signal transduction. Nucleic Acids Research, 48, 3496–3512.
Close, V., Close, W., Kugler, S. J., Reichenzeller, M., Yosifov, D. Y., Bloehdorn, J., Pan, L., Tausch, E., Westhoff, M. A., Dohner, H., Stilgenbauer, S., Oswald, F., & Mertens, D. (2019). FBXW7 mutations reduce binding of NOTCH1, leading to cleaved NOTCH1 accumulation and target gene activation in CLL. Blood, 133, 830–839.
Gianfelici, V. (2012). Activation of the NOTCH1 pathway in chronic lymphocytic leukemia. Haematologica, 97, 328–330.
Zou, Z. J., Fan, L., Wang, L., Xu, J., Zhang, R., Tian, T., Li, J. Y., & Xu, W. (2015). miR-26a and miR-214 down-regulate expression of the PTEN gene in chronic lymphocytic leukemia, but not PTEN mutation or promoter methylation. Oncotarget, 6, 1276–1285.
Zou, Z. J., Zhang, R., Fan, L., Wang, L., Fang, C., Zhang, L. N., Yang, S., Li, Y. Y., Li, J. Y., & Xu, W. (2013). Low expression level of phosphatase and tensin homolog deleted on chromosome ten predicts poor prognosis in chronic lymphocytic leukemia. Leukaemia & Lymphoma, 54, 1159–1164.
Del Papa, B., Baldoni, S., Dorillo, E., De Falco, F., Rompietti, C., Cecchini, D., Cantelmi, M. G., Sorcini, D., Nogarotto, M., Adamo, F. M., Mezzasoma, F., Silva Barcelos, E. C., Albi, E., Iacucci Ostini, R., Di Tommaso, A., Marra, A., Montanaro, G., Martelli, M. P., Falzetti, F., … Sportoletti, P. (2019). Decreased NOTCH1 activation correlates with response to ibrutinib in chronic lymphocytic leukemia. Clinical Cancer Research, 25, 7540–7553.
Rosati, E., Sabatini, R., Rampino, G., Tabilio, A., Di Ianni, M., Fettucciari, K., Bartoli, A., Coaccioli, S., Screpanti, I., & Marconi, P. (2009). Constitutively activated Notch signaling is involved in survival and apoptosis resistance of B-CLL cells. Blood, 113, 856–865.
Weng, A. P., Millholland, J. M., Yashiro-Ohtani, Y., Arcangeli, M. L., Lau, A., Wai, C., Del Bianco, C., Rodriguez, C. G., Sai, H., Tobias, J., Li, Y., Wolfe, M. S., Shachaf, C., Felsher, D., Blacklow, S. C., Pear, W. S., & Aster, J. C. (2006). c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes & Development., 20, 2096–2109.
Tottone, L., Zhdanovskaya, N., Carmona Pestana, A., Zampieri, M., Simeoni, F., Lazzari, S., Ruocco, V., Pelullo, M., Caiafa, P., Felli, M. P., Checquolo, S., Bellavia, D., Talora, C., Screpanti, I., & Palermo, R. (2019). Histone modifications drive aberrant notch3 expression/activity and growth in T-ALL. Frontiers in Oncology, 9, 198.
Palomero, T., Dominguez, M., & Ferrando, A. A. (2008). The role of the PTEN/AKT pathway in NOTCH1-induced leukemia. Cell Cycle, 7, 965–970.
Hwang, K. K., Chen, X., Kozink, D. M., Gustilo, M., Marshall, D. J., Whitesides, J. F., Liao, H. X., Catera, R., Chu, C. C., & Yan, X. J. (2012). Enhanced outgrowth of EBV-transformed chronic lymphocytic leukemia B cells mediated by coculture with macrophage feeder cells. Blood, 119(7), e35-44.
Yan, P., Xue, Z., Li, D., Ni, S., Wang, C., Jin, X., Zhou, D., Li, X., Zhao, X., & Chen, X. (2021). Dysregulated CRTC1-BDNF signaling pathway in the hippocampus contributes to Abeta oligomer-induced long-term synaptic plasticity and memory impairment. Experimental Neurology, 345, 113812.
Nelson, J. D., Denisenko, O., Sova, P., & Bomsztyk, K. (2006). Fast chromatin immunoprecipitation assay. Nucleic Acids Research, 34(1), e2. https://doi.org/10.1093/nar/gnj004
Wirth, M., Doescher, J., Jira, D., Meier, M. A., Piontek, G., Reiter, R., Schlegel, J., & Pickhard, A. (2016). HES1 mRNA expression is associated with survival in sinonasal squamous cell carcinoma. Oral Surgery, Oral Medicine, Oral Pathology, and Oral Radiology, 122, 491–499.
Ahadi, M., Andrici, J., Sioson, L., Sheen, A., Clarkson, A., & Gill, A. J. (2016). Loss of Hes1 expression is associated with poor prognosis in colorectal adenocarcinoma. Human Pathology, 57, 91–97.
Liu, Z., Fu, Q., Fu, H., Wang, Z., Xu, L., An, H., Li, Y., & Xu, J. (2016). A three-molecule score based on Notch pathway predicts poor prognosis in non-metastasis clear cell renal cell carcinoma. Oncotarget, 7, 68559–68570.
Fonseka, L. N., & Tirado, C. A. (2015). C-myc involvement in chronic lymphocytic leukemia (CLL): A molecular and cytogenetic update. Journal of the Association of Genetic Technologists, 41, 176–183.
Bhattacharya, M., Sharma, A. R., Sharma, G., Patra, B. C., Lee, S. S., & Chakraborty, C. (2020). Interaction between miRNAs and signaling cascades of Wnt pathway in chronic lymphocytic leukemia. Journal of Cellular Biochemistry, 121, 4654–4666.
Aljedai, A., Buckle, A. M., Hiwarkar, P., & Syed, F. (2015). Potential role of Notch signalling in CD34+ chronic myeloid leukaemia cells: Cross-talk between Notch and BCR-ABL. PLoS ONE, 10, e0123016.
Nakahara, F., Kitaura, J., Uchida, T., Nishida, C., Togami, K., Inoue, D., Matsukawa, T., Kagiyama, Y., Enomoto, Y., Kawabata, K. C., Chen-Yi, L., Komeno, Y., Izawa, K., Oki, T., Nagae, G., Harada, Y., Harada, H., Otsu, M., Aburatani, H., … Kitamura, T. (2014). Hes1 promotes blast crisis in chronic myelogenous leukemia through MMP-9 upregulation in leukemic cells. Blood, 123, 3932–3942.
Baldoni, S., Sportoletti, P., Del Papa, B., Aureli, P., Dorillo, E., Rosati, E., Ciurnelli, R., Marconi, P., Falzetti, F., & Di Ianni, M. (2013). NOTCH and NF-kappaB interplay in chronic lymphocytic leukemia is independent of genetic lesion. International Journal of Hematology, 98, 153–157.
Baldoni, S., Del Papa, B., Dorillo, E., Aureli, P., De Falco, F., Rompietti, C., Sorcini, D., Varasano, E., Cecchini, D., Zei, T., Di Tommaso, A., Rosati, E., Alexe, G., Roti, G., Stegmaier, K., Di Ianni, M., Falzetti, F., & Sportoletti, P. (2018). Bepridil exhibits anti-leukemic activity associated with NOTCH1 pathway inhibition in chronic lymphocytic leukemia. International Journal of Cancer, 143, 958–970.
Kapoor, I., Li, Y., Sharma, A., Zhu, H., Bodo, J., Xu, W., Hsi, E. D., Hill, B. T., & Almasan, A. (2019). Resistance to BTK inhibition by ibrutinib can be overcome by preventing FOXO3a nuclear export and PI3K/AKT activation in B-cell lymphoid malignancies. Cell Death & Disease, 10, 924.
Bailon, E., Ugarte-Berzal, E., Amigo-Jimenez, I., Van den Steen, P., Opdenakker, G., Garcia-Marco, J. A., & Garcia-Pardo, A. (2014). Overexpression of progelatinase B/proMMP-9 affects migration regulatory pathways and impairs chronic lymphocytic leukemia cell homing to bone marrow and spleen. Journal of Leukocyte Biology, 96, 185–199.
D’Altri, T., Gonzalez, J., Aifantis, I., Espinosa, L., & Bigas, A. (2011). Hes1 expression and CYLD repression are essential events downstream of Notch1 in T-cell leukemia. Cell Cycle, 10, 1031–1036.
Suresh, S., McCallum, L., Crawford, L. J., Lu, W. H., Sharpe, D. J., & Irvine, A. E. (2013). The matricellular protein CCN3 regulates NOTCH1 signalling in chronic myeloid leukaemia. The Journal of Pathology, 231, 378–387.
Saito, N., Hirai, N., Aoki, K., Suzuki, R., Fujita, S., Nakayama, H., Hayashi, M., Ito, K., Sakurai, T., & Iwabuchi, S. (2019). The oncogene addiction switch from NOTCH to PI3K requires simultaneous targeting of NOTCH and PI3K pathway inhibition in glioblastoma. Cancers, 11, 121.
Guo, J., Fu, W., Xiang, M., Zhang, Y., Zhou, K., Xu, C. R., Li, L., Kuang, D., & Ye, F. (2019). Notch1 drives the formation and proliferation of intrahepatic cholangiocarcinoma. Current Medical Science, 39, 929–937.
Hales, E. C., Orr, S. M., Larson Gedman, A., Taub, J. W., & Matherly, L. H. (2013). Notch1 receptor regulates AKT protein activation loop (Thr308) dephosphorylation through modulation of the PP2A phosphatase in phosphatase and tensin homolog (PTEN)-null T-cell acute lymphoblastic leukemia cells. Journal of Biological Chemistry, 288, 22836–22848.
Gao, F., Huang, W., Zhang, Y., Tang, S., Zheng, L., Ma, F., Wang, Y., Tang, H., & Li, X. (2015). Hes1 promotes cell proliferation and migration by activating Bmi-1 and PTEN/Akt/GSK3beta pathway in human colon cancer. Oncotarget, 6, 38667–38680.
Arya, D., Sachithanandan, S. P., Ross, C., Palakodeti, D., Li, S., & Krishna, S. (2017). MiRNA182 regulates percentage of myeloid and erythroid cells in chronic myeloid leukemia. Cell Death Disease, 8, e2547.
Simutis, F. J., Sanderson, T. P., Pilcher, G. D., & Graziano, M. J. (2018). Nonclinical safety assessment of the gamma-secretase inhibitor avagacestat. Toxicological Sciences, 163, 525–542.
Fabbri, G., Holmes, A. B., Viganotti, M., Scuoppo, C., Belver, L., Herranz, D., Yan, X. J., Kieso, Y., Rossi, D., Gaidano, G., Chiorazzi, N., Ferrando, A. A., & Dalla-Favera, R. (2017). Common nonmutational NOTCH1 activation in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the USA, 114, E2911–E2919.
Carra, G., Panuzzo, C., Torti, D., Parvis, G., Crivellaro, S., Familiari, U., Volante, M., Morena, D., Lingua, M. F., Brancaccio, M., Guerrasio, A., Pandolfi, P. P., Saglio, G., Taulli, R., & Morotti, A. (2017). Therapeutic inhibition of USP7-PTEN network in chronic lymphocytic leukemia: A strategy to overcome TP53 mutated/deleted clones. Oncotarget, 8, 35508–35522.
Palacios, F., Prieto, D., Abreu, C., Ruiz, S., Morande, P., Fernandez-Calero, T., Libisch, G., Landoni, A. I., & Oppezzo, P. (2015). Dissecting chronic lymphocytic leukemia microenvironment signals in patients with unmutated disease: MicroRNA-22 regulates phosphatase and tensin homolog/AKT/FOXO1 pathway in proliferative leukemic cells. Leukaemia & Lymphoma, 56, 1560–1565.
Funding
This study was funded by the General items of Health Department of Zhejiang Province (2021KY1078), the Zhejiang Provincial Health Science and Technology Plan in 2022 (Clinical Research Application Project) (2022KY350), and the Wenzhou Science and Technology Bureau (Y20210155).
Author information
Authors and Affiliations
Contributions
CZ and WZ conceived and designed research. WY performed experiments. BL interpreted results of experiments. SW analyzed data. YC prepared figures. QZ performed the bioinformatics analysis and drafted paper. JG, ZZ, and YH edited and revised the manuscript. All authors read and approved final version of manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12033_2022_476_MOESM1_ESM.jpg
Supplementary file1 (DOCX 1499 KB). Supplementary Figure 1. Bioinformatics prediction of HES1-related signaling pathways in CLL. A, HES1-related signaling pathways predicted by the Wikipathways website. B, Protein–protein interaction network of the top 30 differentially expressed genes in the GSE26725 dataset analyzed on STRING.
12033_2022_476_MOESM2_ESM.jpg
Supplementary file2 (DOCX 429 KB). Supplementary Figure 2. The highest expression of HES1 in the MEC1 cell line and the lowest in the HG3 cell line. A, HES1 mRNA expression in CLL-AAT, MEC2, 183-E95, MEC1, and HG3 cell lines determined by RT-qPCR. B, NOTCH1 and PTEN mRNA expression in MEC1 and HG3 cell lines determined by RT-qPCR. *p < 0.05 vs. CLL-AAT cells. #p < 0.05 vs. MEC1 cell line. The experiment was repeated 3 times independently.
Rights and permissions
About this article
Cite this article
Zhang, Q., Zhu, Z., Guan, J. et al. Hes1 Controls Proliferation and Apoptosis in Chronic Lymphoblastic Leukemia Cells by Modulating PTEN Expression. Mol Biotechnol 64, 1419–1430 (2022). https://doi.org/10.1007/s12033-022-00476-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00476-2