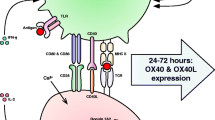
Abstract
CD8 + T cells undergo rapid expansion followed by contraction and the development of memory cells after their receptors are activated. The development of immunological memory following acute infection is a complex phenomenon that involves several molecular, transcriptional, and metabolic mechanisms. As memory cells confer long-term protection and respond to secondary stimulation with strong effector function, understanding the mechanisms that influence their development is of great importance. Orphan nuclear receptors, NR4As, are immediate early genes that function as transcription factors and bind with the NBRE region of chromatin. Interestingly, the NBRE region of activated CD8 + T cells is highly accessible at the same time the expression of NR4As is induced. This suggests a potential role of NR4As in the early events post T cell activation that determines cell fate decisions. In this review, we will discuss the influence of NR4As on the differentiation of CD8 + T cells during the immune response to acute infection and the development of immunological memory. We will also discuss the signals, transcription factors, and metabolic mechanisms that control cell fate decisions.
Highlights
-
Memory CD8 + T cells are an essential subset that mediates long-term protection after pathogen encounters.
-
Some specific environmental cues, transcriptional factors, and metabolic pathways regulate the differentiation of CD8 + T cells and the development of memory cells.
-
Orphan nuclear receptor NR4As are early genes that act as transcription factors and are highly expressed post-T cell receptor activation.
-
NR4As influence the effector function and differentiation of CD8 + T cells and also control the development of immunological memory following acute infection.


Similar content being viewed by others
Data availability
Not applicable.
Abbreviations
- NR4As:
-
Orphan nuclear receptors NR4As
- SLEC:
-
Short-lived effector cells
- MPEC:
-
Memory precursor effector cells
- DARs:
-
Differentially accessible regions
- IL:
-
Interleukin
- INF:
-
Interferon
- CD127:
-
IL-7 receptor alpha subunit
- KLRG1:
-
Killer cell lectin-like receptor G1
- TEM:
-
Effector memory CD8 + T cells
- TCM:
-
Central memory CD8 + T cells
- TRM:
-
Resident memory CD8 + T cells
- LCMV:
-
Lymphocytic choriomeningitis virus
- CTLs:
-
Cytotoxic T-lymphocytes
- TCR:
-
T cell receptor
- mTOR:
-
Mammalian target of rapamycin
- FOXO1:
-
Forkhead box O1
- CD122:
-
IL-2 receptor beta subunit, IL-2Rβ
- BLIMP1:
-
B lymphocyte-induced maturation protein 1
- Bcl-6:
-
B cell lymphoma-6
- STAT:
-
Signal transducer and activator of transcription
- ID2:
-
Inhibitors of DNA binding 2
- ID3:
-
Inhibitors of DNA binding 3
- AP-1:
-
Activator protein-1
- bZIP:
-
Basic leucine zipper proteins
- ATF:
-
Activating transcription factor
- BATF:
-
Basic leucine zipper transcriptional factor ATF-like
- MAF:
-
Musculoaponeurotic fibrosarcoma
- IRF:
-
Interferon regulatory factor
- JAK:
-
Janus kinase
- Tyk:
-
Tyrosine kinase
- SH2:
-
Src homology 2
- SOCS:
-
Suppressors of cytokine signaling
- IgE:
-
Immunoglobulin E
- PI3K:
-
Phosphoinositide 3 kinase
- AKT:
-
Protein kinase B
- mTOR:
-
Mammalian target of rapamycin
- AMPK:
-
5′-Adenosine monophosphate-activated protein kinase
- TRAF6:
-
TNF receptor associated factor 6
- FoxP3:
-
Forkhead box P3
- RORγt:
-
Retinoic acid-related orphan receptor gamma t
- GATA3:
-
GATA binding protein 3
- KLF2:
-
Krüppel-like factor 2
- NFAT:
-
Nuclear factor of activated T cells
- GFP:
-
Green fluorescent protein
- scRNAseq:
-
Single-cell RNA sequencing
- ATAC-Seq:
-
Assay for transposase-accessible chromatin with sequencing
- ATP:
-
Adenosine triphosphate
- MHC:
-
Major histocompatibility complex
- HSV:
-
Herpes simplex virus
- NKT:
-
Natural killer T
References
Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4(12):1191–8. https://doi.org/10.1038/ni1009.
Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27(2):281–95. https://doi.org/10.1016/j.immuni.2007.07.010.
Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med. 2008;205(3):625–40. https://doi.org/10.1084/jem.20071641.
Williams MA, Bevan MJ. Effector and memory CTL differentiation. Annu Rev Immunol. 2007;25:171–92.
Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460(7251):108–12. https://doi.org/10.1038/nature08155.
Mathieu M, Duval F, Daudelin JF, Labrecque N. The Notch signaling pathway controls short-lived effector CD8+ T cell differentiation but is dispensable for memory generation. J Immunol. 2015;194(12):5654–62. https://doi.org/10.4049/jimmunol.1402837.
Roychoudhuri R, Clever D, Li P, Wakabayashi Y, Quinn KM, Klebanoff CA, Ji Y, Sukumar M, Eil RL, Yu Z, Spolski R, Palmer DC, Pan JH, Patel SJ, Macallan DC, Fabozzi G, Shih HY, Kanno Y, Muto A, Zhu J, Gattinoni L, O’Shea JJ, Okkenhaug K, Igarashi K, Leonard WJ, Restifo NP. BACH2 regulates CD8(+) T cell differentiation by controlling access of AP-1 factors to enhancers. Nat Immunol. 2016;17(7):851–60. https://doi.org/10.1038/ni.3441.
Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, Vinup KE, Mrass P, Oliaro J, Killeen N, Orange JS, Russell SM, Weninger W, Reiner SL. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315(5819):1687–91. https://doi.org/10.1126/science.1139393.
Scott-Browne JP, López-Moyado IF, Trifari S, Wong V, Chavez L, Rao A, Pereira RM. Dynamic changes in chromatin accessibility occur in CD8+ T cells responding to viral infection. Immunity. 2016;45(6):1327–40. https://doi.org/10.1016/j.immuni.2016.10.028.
Yu B, Zhang K, Milner JJ, Toma C, Chen R, Scott-Browne JP, Pereira RM, Crotty S, Chang JT, Pipkin ME, Wang W, Goldrath AW. Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat Immunol. 2017;18(5):573–82. https://doi.org/10.1038/ni.3706.
Pearen MA, Muscat GEO. Minireview: nuclear hormone receptor 4A signaling: implications for metabolic disease. Mol Endocrinol. 2010;24:1891–903.
Mognol GP, Spreafico R, Wong V, Scott-Browne JP, Togher S, Hoffmann A, Hogan PG, Rao A, Trifari S. Exhaustion-associated regulatory regions in CD8+ tumor-infiltrating T cells. Proc Natl Acad Sci U S A. 2017;114(13):E2776–85. https://doi.org/10.1073/pnas.1620498114.
Pauken KE, Sammons MA, Odorizzi PM, Manne S, Godec J, Khan O, Drake AM, Chen Z, Sen DR, Kurachi M, Barnitz RA, Bartman C, Bengsch B, Huang AC, Schenkel JM, Vahedi G, Haining WN, Berger SL, Wherry EJ. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 2016;354(6316):1160–5. https://doi.org/10.1126/science.aaf2807.
Boulet S, Daudelin JF, Odagiu L, Pelletier AN, Yun TJ, Lesage S, Cheong C, Labrecque N. The orphan nuclear receptor NR4A3 controls the differentiation of monocyte-derived dendritic cells following microbial stimulation. Proc Natl Acad Sci U S A. 2019;116(30):15150–9. https://doi.org/10.1073/pnas.1821296116.
Wang D, Diao H, Getzler AJ, Rogal W, Frederick MA, Milner J, Yu B, Crotty S, Goldrath AW, Pipkin ME. The transcription factor Runx3 establishes chromatin accessibility of cis-regulatory landscapes that drive memory cytotoxic T lymphocyte formation. Immunity. 2018;48(4):659-674.e6. https://doi.org/10.1016/j.immuni.2018.03.028.
Harty JT, Badovinac VP. Shaping and reshaping CD8+ T-cell memory. Nat Rev Immunol. 2008;8:107–19.
Blanpain C, Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol. 2009;10:207–17.
Ahmed R, Bevan MJ, Reiner SL, Fearon DT. The precursors of memory: models and controversies. Nat Rev Immunol. 2009;9:662–8.
Lefrancois L, Obar JJ. Once a killer, always a killer: from cytotoxic T cell to memory cell. Immunol Rev. 2010;235:206–18.
Bannard O, Kraman M, Fearon DT. Secondary replicative function of CD8+ T cells that had developed an effector phenotype. Science. 2009;323:505–9.
Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, Ahmed R. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol. 2003;4:225–34.
Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol. 2002;2:251–62.
Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27:393–405.
Surh CD, Boyman O, Purton JF, Sprent J. Homeostasis of memory T cells. Immunol Rev. 2006;211:154–63. https://doi.org/10.1111/j.0105-2896.2006.00401.x.
Joshi NS, Kaech SM. Effector CD8 T cell development: a balancing act between memory cell potential and terminal differentiation. J Immunol. 2008;180:1309–15.
Rutishauser RL, Martins GA, Kalachikov S, Chandele A, Parish IA, Meffre E, Jacob J, Calame K, Kaech SM. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31(2):296–308. https://doi.org/10.1016/j.immuni.2009.05.014.
Stemberger C, Huster KM, Koffler M, Anderl F, Schiemann M, Wagner H, Busch DH. A single naive CD8+ T cell precursor can develop into diverse effector and memory subsets. Immunity. 2007;27(6):985–97. https://doi.org/10.1016/j.immuni.2007.10.012.
Schluns KS, Kieper WC, Jameson SC, Lefrançois L. Interleukin-7 mediates the homeostasis of naïve and memory CD8 T cells in vivo. Nat Immunol. 2000;1(5):426–32. https://doi.org/10.1038/80868.
Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401(6754):708–12. https://doi.org/10.1038/44385.
Rubinstein MP, Lind NA, Purton JF, Filippou P, Best JA, McGhee PA, Surh CD, Goldrath AW. IL-7 and IL-15 differentially regulate CD8+ T-cell subsets during contraction of the immune response. Blood. 2008;112(9):3704–12. https://doi.org/10.1182/blood-2008-06-160945.
Obar JJ, Molloy MJ, Jellison ER, Stoklasek TA, Zhang W, Usherwood EJ, Lefrançois L. CD4+ T cell regulation of CD25 expression controls development of short-lived effector CD8+ T cells in primary and secondary responses. Proc Natl Acad Sci U S A. 2010;107(1):193–8. https://doi.org/10.1073/pnas.0909945107.
Jabbari A, Harty JT. Secondary memory CD8+ T cells are more protective but slower to acquire a central-memory phenotype. J Exp Med. 2006;203:919–32.
Masopust D, Ha SJ, Vezys V, Ahmed R. Stimulation history dictates memory CD8 T cell phenotype: implications for prime-boost vaccination. J Immunol. 2006;177(2):831–9. https://doi.org/10.4049/jimmunol.177.2.831.
Nolz JC, Harty JT. Protective capacity of memory CD8+ T cells is dictated by antigen exposure history and nature of the infection. Immunity. 2011;34:781–93.
Joshi NS, Cui W, Dominguez CX, Chen JH, Hand TW, Kaech SM. Increased numbers of preexisting memory CD8 T cells and decreased T-bet expression can restrain terminal differentiation of secondary effector and memory CD8 T cells. J Immunol. 2011;187(8):4068–76. https://doi.org/10.4049/jimmunol.1002145.
van der Windt GJ, Everts B, Chang CH, Curtis JD, Freitas TC, Amiel E, Pearce EJ, Pearce EL. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36(1):68–78. https://doi.org/10.1016/j.immuni.2011.12.007.
Xue HH, Zhao DM. Regulation of mature T cell responses by the Wnt signaling pathway. Ann N Y Acad Sci. 2012;1247:16–33. https://doi.org/10.1111/j.1749-6632.2011.06302.x.
Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity. 2010;33(2):229–40. https://doi.org/10.1016/j.immuni.2010.08.002.
Schenkel JM, Fraser KA, Vezys V, Masopust D. Sensing and alarm function of resident memory CD8+ T cells. Nat Immunol. 2013;14:509–13.
Schenkel JM, Fraser KA, Beura LK, Pauken KE, Vezys V, Masopust D. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science. 2014;346:98–101.
Gebhardt T, Wakim LM, Eidsmo L, Reading PC, Heath WR, Carbone FR. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol. 2009;10:524–30.
Pan Y, Tian T, Park CO, Lofftus SY, Mei S, Liu X, Luo C, O’Malley JT, Gehad A, Teague JE. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature. 2017;543:252–6.
Steinbach K, Vincenti I, Kreutzfeldt M, Page N, Muschaweckh A, Wagner I, Drexler I, Pinschewer D, Korn T, Merkler D. Brain-resident memory T cells represent an autonomous cytotoxic barrier to viral infection. J Exp Med. 2016;213:1571–87.
Mackay LK, Wynne-Jones E, Freestone D, Pellicci DG, Mielke LA, Newman DM, Braun A, Masson F, Kallies A, Belz GT, Carbone FR. T-box transcription factors combine with the cytokines TGF-β and IL-15 to control tissue-resident memory T cell fate. Immunity. 2015;43(6):1101–11. https://doi.org/10.1016/j.immuni.2015.11.008.
Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon ML, Vega-Ramos J, Lauzurica P, Mueller SN, Stefanovic T, Tscharke DC, Heath WR, Inouye M, Carbone FR, Gebhardt T. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat Immunol. 2013;12:1294–301. https://doi.org/10.1038/ni.2744.
Mackay LK, Minnich M, Kragten NA, Liao Y, Nota B, Seillet C, Zaid A, Man K, Preston S, Freestone D, Braun A, Wynne-Jones E, Behr FM, Stark R, Pellicci DG, et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 2016;352(6284):459–63. https://doi.org/10.1126/science.aad2035.
Wakim LM, Woodward-Davis A, Liu R, Hu Y, Villadangos J, Smyth G, Bevan MJ. The molecular signature of tissue resident memory CD8 T cells isolated from the brain. J Immunol. 2012;189(7):3462–71. https://doi.org/10.4049/jimmunol.1201305.
Stelma F, de Niet A, Sinnige MJ, van Dort KA, van Gisbergen KPJM, Verheij J. Human intrahepatic CD69+ CD8+ T cells have a tissue resident memory T cell phenotype with reduced cytolytic capacity. Sci Rep. 2017;7(1):6172. https://doi.org/10.1038/s41598-017-06352-3.
Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, Senda T, Sun X, Ho SH, Lerner H, Friedman AL, Shen Y, Farber DL. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 2017;20(12):2921–34. https://doi.org/10.1016/j.celrep.2017.08.078.
Pallett LJ, Davies J, Colbeck EJ, Robertson F, Hansi N, Easom NJW, Burton AR, Stegmann KA, Schurich A, Swadling L, Gill US, Male V, Luong T, Gander A, Davidson BR, Kennedy PTF, Maini MK. IL-2high tissue-resident T cells in the human liver: sentinels for hepatotropic infection. J Exp Med. 2017;214(6):1567–80. https://doi.org/10.1084/jem.20162115.
Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, Wang D, Getzler AJ, Nguyen T, Crotty S, Wang W, Pipkin ME, Goldrath AW. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature. 2017;552(7684):253–7. https://doi.org/10.1038/nature24993.
Ebihara T, Song C, Ryu SH, Plougastel-Douglas B, Yang L, Levanon D, Groner Y, Bern MD, Stappenbeck TS, Colonna M. Runx3 specifies lineage commitment of innate lymphoid cells. Nat Immunol. 2015;16:1124–33.
Rapp M, Lau CM, Adams NM, Weizman OE, O’Sullivan TE, Geary CD, Sun JC. Core-binding factor β and Runx transcription factors promote adaptive natural killer cell responses. Sci Immunol. 2017;2(18):eaan3796. https://doi.org/10.1126/sciimmunol.aan3796.
Grueter B, Petter M, Egawa T, Laule-Kilian K, Aldrian CJ, Wuerch A, Ludwig Y, Fukuyama H, Wardemann H, Waldschuetz R, Möröy T, Taniuchi I, Steimle V, Littman DR, Ehlers M. Runx3 regulates integrin alpha E/CD103 and CD4 expression during development of CD4-/CD8+ T cells. J Immunol. 2005;175(3):1694–705. https://doi.org/10.4049/jimmunol.175.3.1694.
Reis BS, Rogoz A, Costa-Pinto FA, Taniuchi I, Mucida D. Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4+ T cell immunity. Nat Immunol. 2013;14:271–80.
Reis BS, Hoytema van Konijenburg DP, Grivennikov SI, Mucida D. Transcription factor T-bet regulates intraepithelial lymphocyte functional maturation. Immunity. 2014;41:244–56.
Laidlaw BJ, Zhang N, Marshall HD, Staron MM, Guan T, Hu Y, Cauley LS, Craft J, Kaech SM. CD4+ T cell help guides formation of CD103+ lung-resident memory CD8+ T cells during influenza viral infection. Immunity. 2014;41:633–45.
Hendriks J, Xiao Y, Rossen JW, van der Sluijs KF, Sugamura K, Ishii N, Borst J. During viral infection of the respiratory tract, CD27, 4–1BB, and OX40 collectively determine formation of CD8+ memory T cells and their capacity for secondary expansion. J Immunol. 2005;175(3):1665–76. https://doi.org/10.4049/jimmunol.175.3.1665.
Pulle G, Vidric M, Watts TH. IL-15-dependent induction of 4–1BB promotes antigen-independent CD8 memory T cell survival. J Immunol. 2006;176:2739–48.
Obar JJ, Jellison ER, Sheridan BS, Blair DA, Pham QM, Zickovich JM, Lefrançois L. Pathogen-induced inflammatory environment controls effector and memory CD8+ T cell differentiation. J Immunol. 2011;187(10):4967–78. https://doi.org/10.4049/jimmunol.1102335.
Mescher MF, Curtsinger JM, Agarwal P, Casey KA, Gerner M, Hammerbeck CD, Popescu F, Xiao Z. Signals required for programming effector and memory development by CD8+ T cells. Immunol Rev. 2006;211:81–92. https://doi.org/10.1111/j.0105-2896.2006.00382.x.
Curtsinger JM, Johnson CM, Mescher MF. CD8 T cell clonal expansion and development of effector function require prolonged exposure to antigen, costimulation, and signal 3 cytokine. J Immunol. 2003;171:5165–71.
Kolumam GA, Thomas S, Thompson LJ, Sprent J, Murali-Krishna K. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J Exp Med. 2005;202:637–50.
Whitmire JK, Eam B, Benning N, Whitton JL. Direct interferon-γ signaling dramatically enhances CD4+ and CD8+ T cell memory. J Immunol. 2007;179:1190–7.
Agarwal P, Raghavan A, Nandiwada SL, Curtsinger JM, Bohjanen PR, Mueller DL, Mescher MF. Gene regulation and chromatin remodeling by IL-12 and type I IFN in programming for CD8 T cell effector function and memory. J Immunol. 2009;183(3):1695–704. https://doi.org/10.4049/jimmunol.0900592.
Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458:211–4.
Kaech SM, Ahmed R. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naive cells. Nature Immunol. 2001;2:415–22.
D’Souza WN, Hedrick SM. Cutting edge: latecomer CD8 T cells are imprinted with a unique differentiation program. J Immunol. 2006;177:777–81.
Cui W, Joshi NS, Jiang A, Kaech SM. Effects of signal 3 during CD8 T cell priming: bystander production of IL-12 enhances effector T cell expansion but promotes terminal differentiation. Vaccine. 2009;27:2177–87.
Pearce EL, Shen H. Generation of CD8 T cell memory is regulated by IL-12. J Immunol. 2007;179:2074–81.
Wiesel M, Crouse J, Bedenikovic G, Sutherland A, Joller N, Oxenius A. Type-I IFN drives the differentiation of short-lived effector CD8+ T cells in vivo. Eur J Immunol. 2012;42(2):320–9. https://doi.org/10.1002/eji.201142091.
Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32:79–90.
Intlekofer AM, Takemoto N, Kao C, Banerjee A, Schambach F, Northrop JK, Shen H, Wherry EJ, Reiner SL. Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J Exp Med. 2007;204(9):2015–21. https://doi.org/10.1084/jem.20070841
Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, Wherry JE, Reiner SL. Cutting edge: the transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol. 2010;185:4988–92.
Intlekofer AM, Banerjee A, Takemoto N, Gordon SM, Dejong CS, Shin H, Hunter CA, Wherry EJ, Lindsten T, Reiner SL. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 2008;321:408–11.
Takemoto N, Intlekofer AM, Northrup JT, Wherry EJ, Reiner SL. Cutting edge: IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J Immunol. 2006;177:7515–9.
Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and eomesodermin. Immunity. 2010;32:67–78.
Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. NatureImmunol. 2010;11:114–20.
Kwon H, Thierrt-Meg D, Thierry-Mieg J, Kim HP, Oh J, Tunyaplin C, Carotta S, Donovan CE, Goldman ML, Tailor P, Ozato K, Levy DE, Nutt SL, Calame K, Leonard WJ. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 2009;31:941–52.
Shin H, Blackburn SD, Intlekofer AM, Kao C, Angelosanto JM, Reiner SL, Wherry JE. A role for the transcriptional repressor Blimp-1 in CD8+ T cell exhaustion during chronic viral infection. Immunity. 2009;31:309–20.
Cui W, Liu Y, Weinstein JS, Craft J, Kaech SM. An interleukin-21–interleukin-10–STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity. 2011;35:792–805.
Ichii H, Sakamoto A, Arima M, Hatano M, Kuroda Y, Tokuhisa T. Bcl6 is essential for the generation of long-term memory CD4+ T cells. Int Immunol. 2007;19:427–33.
Ji Y, Pos Z, Rao M, Klebanoff CA, Yu Z, Sukumar M, Reger RN, Palmer DC, Borman ZA, Muranski P, Wang E, Schrump DS, Marincola FM, Restifo NP, Gattinoni L. Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat Immunol. 2011;12(12):1230–7. https://doi.org/10.1038/ni.2153.
Cannarile MA, Lind NA, Rivera R, Sheridan AD, Camfield KA, Wu BB, Cheung KP, Ding Z, Goldrath AW. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nature Immunol. 2006;7:1317–25.
Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, Li HS, Rivera RR, Lind KC, D’Cruz LM, Watowich SS, Murre C, Goldrath AW. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol. 2011;12(12):1221–9. https://doi.org/10.1038/ni.2158.
Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;11:859–68. https://doi.org/10.1038/nrc1209.
Milde-Langosch K. The Fos family of transcription factors and their role in tumourigenesis. Eur J Cancer. 2005;16:2449–61. https://doi.org/10.1016/j.ejca.2005.08.008.
Hernandez JM, Floyd DH, Weilbaecher KN, Green PL, Boris-Lawrie K. Multiple facets of junD gene expression are atypical among AP-1 family members. Oncogene. 2008;7(35):4757–67. https://doi.org/10.1038/onc.2008.120.
Murphy TL, Tussiwand R, Murphy KM. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat Rev Immunol. 2013;13(7):499–509. https://doi.org/10.1038/nri3470.
Quigley M, Pereyra F, Nilsson B, Porichis F, Fonseca C, Eichbaum Q, Julg B, Jesneck JL, Brosnahan K, Imam S, Russell K, Toth I, Piechocka-Trocha A, et al. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat Med. 2010;16(10):1147–51. https://doi.org/10.1038/nm.2232.
Kuroda S, Yamazaki M, Abe M, Sakimura K, Takayanagi H, Iwai Y. Basic leucine zipper transcription factor, ATF-like (BATF) regulates epigenetically and energetically effector CD8 T-cell differentiation via Sirt1 expression. Proc Natl Acad Sci U S A. 2011;108(36):14885–9. https://doi.org/10.1073/pnas.1105133108.
Kurachi M, Barnitz RA, Yosef N, Odorizzi PM, DiIorio MA, Lemieux ME, Yates K, Godec J, Klatt MG, Regev A, Wherry EJ, Haining WN. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat Immunol. 2014;15(4):373–83. https://doi.org/10.1038/ni.2834.
Logan MR, Jordan-Williams KL, Poston S, Liao J, Taparowsky EJ. Overexpression ofBATFf induces an apoptotic defect and an associated lymphoproliferative disorder in mice. Cell Death Dis. 2012;3:e310.
Man K, Gabriel SS, Liao Y, Gloury R, Preston S, Henstridge DC, Pellegrini M, Zehn D, Berberich-Siebelt F, Febbraio MA, Shi W, Kallies A. Transcription factor IRF4 promotes CD8+ T cell exhaustion and limits the development of memory-like T cells during chronic infection. Immunity. 2017;47(6):1129-1141.e5. https://doi.org/10.1016/j.immuni.2017.11.021.
Lim CP, Cao X. Structure, function, and regulation of STAT proteins. Mol BioSyst. 2006;2(11):536–50.
Braunstein J, Brutsaert S, Olson R, Schindler C. STATs dimerize in the absence of phosphorylation. J Biol Chem. 2003;278(36):34133–40. https://doi.org/10.1074/jbc.M304531200.
Darnell JE Jr. STATs and gene regulation. Science. 1997;277(5332):1630–5. https://doi.org/10.1126/science.277.5332.1630.
Ihle JN, Witthuhn BA, Quelle FW, Yamamoto K, Silvennoinen O. Signaling through the hematopoietic cytokine receptors. Annu Rev Immunol. 1995;13:369–98. https://doi.org/10.1146/annurev.iy.13.040195.002101.
Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, King KL, Sheehan KC, Yin L, Pennica D, Johnson EM Jr, Schreiber RD. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell. 1998;93(3):373–83. https://doi.org/10.1016/s0092-8674(00)81166-6.
Nosaka T, van Deursen JM, Tripp RA, Thierfelder WE, Witthuhn BA, McMickle AP, Doherty PC, Grosveld GC, Ihle JN. Defective lymphoid development in mice lacking Jak3. Science. 1995;270(5237):800–2. https://doi.org/10.1126/science.270.5237.800.
Spolski R, Li P, Leonard WJ. Biology and regulation of IL-2: from molecular mechanisms to human therapy. Nat Rev Immunol. 2018;18(10):648–59. https://doi.org/10.1038/s41577-018-0046-y.
Siegel AM, Heimall J, Freeman AF, Hsu AP, Brittain E, Brenchley JM, Douek DC, Fahle GH, Cohen JI, Holland SM, Milner JD. A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity. 2011;35(5):806–18. https://doi.org/10.1016/j.immuni.2011.09.016.
Seki Y, Hayashi K, Matsumoto A, Seki N, Tsukada J, Ransom J, Naka T, Kishimoto T, Yoshimura A, Kubo M. Expression of the suppressor of cytokine signaling-5 (SOCS5) negatively regulates IL-4-dependent STAT6 activation and Th2 differentiation. Proc Natl Acad Sci U S A. 2002;99(20):13003–8. https://doi.org/10.1073/pnas.202477099.
Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–84.
Lohoff M, Mak TW. Roles of interferon regulatory factors in T-helper-cell differentiation. Nat Rev Immunol. 2005;5:125–35.
Pernis AB. The role of IRF-4 in B and T cell activation and differentiation. J Interferon Cytokine Res. 2002;22:111–20.
Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7(7):773–82. https://doi.org/10.1038/ni1357.
Xu WD, Pan HF, Ye DQ, Xu Y. Targeting IRF4 in autoimmune diseases. Autoimmun Rev. 2012;11:918–24.
Brüstle A, Heink S, Huber M, Rosenplänter C, Stadelmann C, Yu P, Arpaia E, Mak TW, Kamradt T, Lohoff M. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8(9):958–66. https://doi.org/10.1038/ni1500.
Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkhurst CN, Muratet M, Newberry KM, Meadows S, Greenfield A, Yang Y, Jain P, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151(2):289–303. https://doi.org/10.1016/j.cell.2012.09.016.
Koch S, Mousset S, Graser A, Reppert S, Übel C, Reinhardt C, Zimmermann T, Rieker R, Lehr HA, Finotto S. IL-6 activated integrated BATF/IRF4 functions in lymphocytes are T-bet-independent and reversed by subcutaneous immunotherapy. Sci Rep. 2013;3:1754. https://doi.org/10.1038/srep01754.
Man K, Miasari M, Shi W, Xin A, Henstridge DC, Preston S, Pellegrini M, Belz GT, Smyth GK, Febbraio MA, et al. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat Immunol. 2013;14:1155–65.
Yao S, Buzo BF, Pham D, Jiang L, Taparowsky EJ, Kaplan MH, Sun J. Interferon regulatory factor 4 sustains CD8(+) T cell expansion and effector differentiation. Immunity. 2013;39:833–45.
Raczkowski F, Ritter J, Heesch K, Schumacher V, Guralnik A, Ho¨cker L, Raifer H, Klein M, Bopp T, Harb H, et al. The transcription factor interferon regulatory factor 4 is required for the generation of protective effector CD8+ T cells. Proc Natl Acad Sci USA. 2013;110:15019–15024.
Wang R, Green DR. Metabolic checkpoints in activated T cells. Nat Immunol. 2012;13:907–15.
Winoto A, Littman DR. Nuclear hormone receptors in T lymphocytes. Cell. 2002;109(Suppl.):S57–S66.
Li Q, Ke XN, Sundaram R, Wong-Staal F. NR4A1, 2, 3–an orphan nuclear hormone receptor family involved in cell apoptosis and carcinogenesis. Histol Histopathol. 2006;21:533–40.
Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, Hogquist KA. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208(6):1279–89. https://doi.org/10.1084/jem.20110308.
Bending D, Prieto Martín P, Paduraru A, Ducker C, Marzaganov E, Laviron M, Kitano S, Miyachi H, Crompton T, Ono M. A timer for analyzing temporally dynamic changes in transcription during differentiation in vivo. J Cell Biol. 2018;217(8):2931–50. https://doi.org/10.1083/jcb.201711048.
Richard AC, Lun ATL, Lau WWY, Göttgens B, Marioni JC, Griffiths GM. T cell cytolytic capacity is independent of initial stimulation strength. Nat Immunol. 2018;19:849–58. https://doi.org/10.1038/s41590-018-0160-9.
Ashouri JF, Weiss A. Endogenous Nur77 is a specific indicator of antigen receptor signaling in human T and B cells. J Immunol. 2017;198:657–68. https://doi.org/10.4049/jimmunol.1601301.
Liu Z, Smith GSW, McLaughlin KA, Schwartz LM, Osborne BA. Apoptotic signals delivered through the T-cell receptor of a T-cell hybrid require the immediate-early gene nur77. Nature. 1994;367:281–4.
Sekiya T, Kashiwagi I, Yoshida R, Fukaya T, Morita R, Kimura A, Ichinose H, Metzger D, Chambon P, Yoshimura A. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat Immunol. 2013;14:230–7.
Best JA, Blair DA, Knell J, Yang E, Mayya V, Doedens A, Dustin ML, Goldrath AW. Immunological genome project consortium. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat Immunol. 2013;14(4):404–12. https://doi.org/10.1038/ni.2536.
Hu QN, Suen AYW, Henao-Caviedes LM, Baldwin TA. Nur77 regulates nondeletional mechanisms of tolerance in T cells. J Immunol. 2017;199:3147–57.
Liu X, Wang Y, Lu H, Li J, Yan X, Xiao M, Hao J, Alekseev A, Khong H, Chen T, Huang R, Wu J, Zhao Q, et al. Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature. 2019;567(7749):525–9. https://doi.org/10.1038/s41586-019-0979-8.
Odagiu L, Boulet S, Maurice De Sousa D, Daudelin J-F, Nicolas S, Labrecque N. Early programming of CD8+ T cell response by the orphan nuclear receptor NR4A3. Proc Natl Acad Sci USA. 2020;117:24392–402. https://doi.org/10.1073/pnas.2007224117.
Nowyhed HN, Huynh TR, Blatchley A, Wu R, Thomas GD, Hedrick CC. The nuclear receptor nr4a1 controls CD8 T cell development through transcriptional suppression of runx3. Sci Rep. 2015;5:9059. https://doi.org/10.1038/srep09059.
Nayar R, Enos M, Prince A, Shin H, Hemmers S, Jiang JK, Klein U, Thomas CJ, Berg LJ. TCR signaling via Tec kinase ITK and interferon regulatory factor 4 (IRF4) regulates CD8+ T-cell differentiation. Proc Natl Acad Sci USA. 2012;109:E2794–802.
Nayar R, Schutten E, Bautista B, Daniels K, Prince AL, Enos M, Brehm MA, Swain SL, Welsh RM, Berg LJ. Graded levels of IRF4 regulate CD8+ T cell differentiation and expansion, but not attrition, in response to acute virus infection. J Immunol. 2014;192:5881–93.
Nowyhed HN, Huynh TR, Thomas GD, Blatchley A, Hedrick CC. Cutting edge: the orphan nuclear receptor Nr4a1 regulates CD8+ T cell expansion and effector function through direct repression of Irf4. J Immunol. 2015;195:3515–9. https://doi.org/10.4049/jimmunol.1403027.
Saijo K, Winner B, Carson CT, Collier JG, Boyer L, Rosenfeld MG, Gage FH, Glass CK. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137(1):47–59. https://doi.org/10.1016/j.cell.2009.01.038.
Boddupalli CS, Nair S, Gray SM, Nowyhed HN, Verma R, Gibson JA, Abraham C, Narayan D, Vasquez J, Hedrick CC, Flavell RA, Dhodapkar KM, Kaech SM, Dhodapkar MV. ABC transporters and NR4A1 identify a quiescent subset of tissue-resident memory T cells. J Clin Invest. 2016;126(10):3905–16. https://doi.org/10.1172/JCI85329.
Kurd NS, He Z, Louis TL, Milner JJ, Omilusik KD, Jin W, Tsai MS, Widjaja CE, Kanbar JN, Olvera JG, Tysl T, Quezada LK, Boland BS, Huang WJ, et al. Early precursors and molecular determinants of tissue-resident memory CD8+ T lymphocytes revealed by single-cell RNA sequencing. Sci Immunol. 2020;5(47):eaaz6894. https://doi.org/10.1126/sciimmunol.aaz6894.
Author information
Authors and Affiliations
Contributions
OOO: conceptualization. OOO, BOA, SPA, JAG, and MS: wrote the manuscript draft. OOO and SPA: review and editing. OOO, BOA, SPA, JAG, and MS: proofreading. All authors agree to the final submission of this manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
Not applicable.
Consent for publication
All authors agree to the final manuscript.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Oladipo, O.O., Adedeji, B.O., Adedokun, S.P. et al. Regulation of effector and memory CD8 + T cell differentiation: a focus on orphan nuclear receptor NR4A family, transcription factor, and metabolism. Immunol Res 71, 314–327 (2023). https://doi.org/10.1007/s12026-022-09353-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12026-022-09353-1