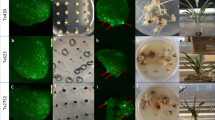
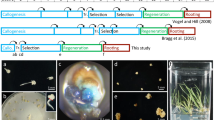
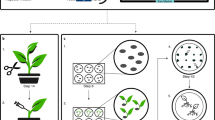
Abstract
The standard stalwart Agrobacterium-mediated transformation protocols for sorghum use differentiating embryogenic callus induced from immature embryos in the genotype BTx430. While reliable, the standard protocols are lengthy, and both time and resource expensive. We have investigated the use of altruistic morphogenic regulator mediated transformation (MRMT) to improve sorghum transformation for use in genome editing. Here, we show a method for rapid transformation and plant regeneration of sorghum (Sorghum bicolor L. cultivar BTx430) mediated by altruistic Baby boom and Wuschel2. Confirming and extending previous recent work using altruistic MRMT, we show that the altruistic MRMT rapid protocol results in a higher transformation frequency expressed as a function of the number of independent events per immature embryos infected and requires less time in a transformation pipeline which translates to an increased number of constructs one full-time equivalent (FTE) can manage per yr. This approach is compatible with genome editing requirements and should help address the transformation bottleneck for functional genomics and genome editing studies in sorghum.








Similar content being viewed by others
References
Able JA, Rathus C, Godwin ID (2001) The investigation of optimal bombardment parameters for transient and stable transgene expression in sorghum. In Vitro Cell Dev Biol - Plant 37:341–348
Altpeter F, Springer NM, Bartley LE, Blechl A, Brutnell TP, Citovsky V, Conrad L, Gelvin SB, Jackson D, Kausch AP, Lemaux PG, Medford JI, Orozo-Cardenas M, Tricoli D, VanEck J, Voytas DF, Walbot V, Wang K, Zhang ZJ, Stewart CN (2016) Advancing crop transformation in the era of genome editing. Plant Cell 28:1510–1520
Anand A, Bass SH, Wu E, Wang N, McBride KE, Annaluru N, Miller M, Hua M, Jones TJ (2018) An improved ternary vector system for Agrobacterium-mediated rapid maize transformation. Plant Mol Biol 97:187–200
Aregawi K, Shen J, Pierroz G, Bucheli C, Sharma M, Dahlberg J, Owiti J, Lemaux PG (2020) Pathway to validate gene function in key bioenergy crop, Sorghum bicolor. bioRxiv 2020.12.08.416347; https://doi.org/10.1101/2020.12.08.416347
Belide S, Vanhercke T, Petrie JR, Singh SP (2017) Robust genetic transformation of sorghum (Sorghum bicolor L.) using differentiating embryogenic callus induced from immature embryos. Plant Methods 13:1–12
Belton PS, Taylor JR (2004) Sorghum and millets: protein sources for Africa. Trends Food Sci Technol 15:94–98
Borisjuk N, Kishchenko O, Eliby S, Schramm C, Anderson P, Jatayev S, Kurishbayev A, Shavrukov Y (2019) Genetic modification for wheat improvement: from transgenesis to genome editing. BioMed Res Int Article ID 6216304 https://doi.org/10.1155/2019/6216304
Casas AM, Kononowicz AK, Zehr UB, Tomes DT, Axtell JD, Butler LG, Bressan RA, Hasegawa PM (1993) Transgenic sorghum plants via microprojectile bombardment. Proc Nat Acad Sci USA 90:11212–11216
Che P, Anand A, Wu E, Sander JD, Simon MK, Zhu W, Sigmund AL, Zastrow-Hayes G, Miller M, Liu D, Lawit SJ, Zhao Z-Y, Albertsen MC, Jones TJ (2018) Developing a flexible, high-efficiency Agrobacterium-mediated sorghum transformation system with broad application. Plant Biotechnol J 16:1388–1395
Che P, Wu E, Simon MK, Anand A, Lowe K, Gao H, Sigmund AL, Yang M, Albertsen MC, Gordon-Kamm W (2021) Wuschel2 enables highly efficient CRISPR/Cas-targeted genome editing during rapid de novo shoot regeneration in sorghum. bioRxiv. bioRxiv 2021.06.21.449302; https://doi.org/10.1101/2021.06.21.449302
Chu UC, Adelberg J, Lowe K, Jones TJ (2019) Use of DoE methodology to optimize the regeneration of high-quality, single-copy transgenic Zea mays L. (maize) plants. In Vitro Cell Dev Biol - Plant 55:678–694
Cooper EA, Brenton ZW, Flinn BS, Jenkins J, Shu S, Flowers D, Luo F, Wang Y, Xia P, Barry K, Daum C, Lipzen A, Yoshinaga Y, Schmutz J, Saski C, Vermerris W, Kresovich S (2019) A new reference genome for Sorghum bicolor reveals high levels of sequence similarity between sweet and grain genotypes: implications for the genetics of sugar metabolism. BMC Genomics 20:1–13
Do PT, Lee H, Nelson-Vasilchik K, Kausch A, Zhang ZJ (2018) Rapid and efficient genetic transformation of sorghum via Agrobacterium-mediated method. Curr Protoc Plant Biol 3:e20077
Duodu KG, Taylor JRN, Belton PS, Hamaker BR (2003) Factors affecting sorghum protein digestibility. J Cereal Sci 38:117–131
Gao Z, Xie X, Ling Y, Muthukrishnan S, Liang GH (2005) Agrobacterium tumefaciens-mediated sorghum transformation using a mannose selection system. Plant Biotechnol J 3:591–599
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2011) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40:D1178–D1186
Grootboom AW, Mkhonza N, O’Kennedy MM, Chakauya E, Kunert KJ, Chikwamba RK (2010) Biolistic mediated sorghum (Sorghum bicolor L. Moench) transformation via mannose and bialaphos based selection systems. Intl J Bot 6:89–94
Gurel S, Gurel E, Kaur R, Wong J, Meng L, Tan H-Q, Lemaux PG (2009) Efficient, reproducible Agrobacterium-mediated transformation of sorghum using heat treatment of immature embryos. Plant Cell Rep 28:429–444
Hague J, Nelson K, Yonchak A, Kausch AP (2022) qPCR methods for the quantification of transgene insert copy number and zygosity using the comparative Ct method in transgenic sorghum bicolor L Moench In: Bilichak A, Laurie JD (eds) Accelerated breeding of cereal crops. Springer Protocols Handbooks, New York pp 1–5
Hao H, Li Z, Leng C, Lu C, Luo H, Liu Y, Wu X, Liu Z, Shang L, Jing H-C (2021) Sorghum breeding in the genomic era: opportunities and challenges. Theor Appl Genet 134:1899–1924
Hensel G (2020) Genetic transformation of Triticeae cereals–summary of almost three-decade’s development. Biotechnol Adv 40:1–8
Hiei Y, Komari T (2008) Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nature Protoc 3:824–834
Hoerster G, Wang N, Ryan L, Wu E, Anand A, McBride K, Lowe K, Jones T, Gordon-Kamm B (2020) Use of non-integrating Zm-Wus2 vectors to enhance maize transformation. In Vitro Cell Dev Biol - Plant 56:265–279
Ibrahim AS, El-Shihy OM, Fahmy AH (2010) Highly efficient Agrobacterium tumefaciens-mediated transformation of elite Egyptian barley cultivars. Am-Eurasian J Sustain Agric 4:403–413
Ishida Y, Hiei Y, Komari T (2007) Agrobacterium-Mediated Transformation of Maize Nature Protoc 2:1614–1621
Kausch AP, Nelson-Vasilchik K, Hague J, Mookkan M, Quemada H, Dellaporta S, Fragoso C, Zhang ZJ (2019) Edit at will: genotype independent plant transformation in the era of advanced genomics and genome editing. Plant Sci 281:186–205
Kausch AP, Nelson-Vasilchik K, Tilelli M, Hague JP (2021a) Maize tissue culture, transformation, and genome editing. In Vitro Cell Dev Biol - Plant 57:653–671
Kausch AP, Wang K, Kaeppler HF, Gordon-Kamm W (2021b) Maize transformation: history, progress, and perspectives. Mol Breed 41:1–36
Kumar AA, Reddy BV, Ramaiah B, Sahrawat KL, Pfeiffer WH (2013) Gene effects and heterosis for grain iron and zinc concentration in sorghum [Sorghum bicolor (L.) Moench]. Field Crops Res 146:86–95
Li A, Jia S, Yobi A, Ge Z, Sato SJ, Zhang C, Angelovici R, Clemente TE, Holding DR (2018) Editing of an alpha-Kafirin gene family increases, digestibility and protein quality in sorghum. Plant Physiol 177:1425–1438
Lin C-Y, Eudes A (2020) Strategies for the production of biochemicals in bioenergy crops. Biotechnol Biofuels 13:1–25
Lin C-Y, Tian Y, Nelson-Vasilchik K, Hague J, Kakumanu R, Lee MY, Trinh J, Northen T, Baidoo E, Kausch A (2021) Engineering sorghum for higher 4-hydroxybenzoic acid content. bioRxiv 2021.07.13.452095; https://doi.org/10.1101/2021.07.13.452095
Lowe K, La Rota M, Hoerster G, Hastings C, Wang N, Chamberlin M, Wu E, Jones T, Gordon-Kamm W (2018) Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell Dev Biol - Plant 54:240–252
Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho M-J, Scelonge C, Lenderts B, Chamberlin M, Cushatt J, Wang L, Ryan L, Khan T, Chow-Yiu J, Hua W, Yu M, Banh J, Bao Z, Brink K, Igo E, Rudrappa B, Shamseer PM, Bruce W, Newman L, Shen B, Zheng P, Bidney D, Falco SC, RegisterIII JC, Zhao Z-Y, Xu D, Jones TJ, Gordon-Kamm WJ (2016) Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28:1998–2015
Mann DG, King ZR, Liu W, Joyce BL, Percifield RJ, Hawkins JS, LaFayette PR, Artelt BJ, Burris JN, Mazarei M, Bennetzen JL, Parrott WA, Stewart CN Jr (2011) Switchgrass (Panicum virgatum L.) polyubiquitin gene (PvUbi1 and PvUbi2) promoters for use in plant transformation. BMC Biotechnol 11:1–14
Mookkan M, Nelson-Vasilchik K, Hague J, Kausch A, Zhang ZJ (2018) Morphogenic regulator-mediated transformation of maize inbred B73. Curr Protoc Plant Biol 3:e20075
Mullet J, Morishige D, McCormick R, Truong S, Hilley J, McKinley B, Anderson R, Olson SN, Rooney W (2014) Energy Sorghum—a genetic model for the design of C4 grass bioenergy crops. J Exp Bot 65:3479–3489
Nelson-Vasilchik K, Hague J, Mookkan M, Zhang ZJ, Kausch A (2018) Transformation of recalcitrant sorghum varieties facilitated by baby boom and Wuschel2. Curr Protoc Plant Biol 3:e20076
Nguyen T-V, Thu TT, Claeys M, Angenon G (2007) Agrobacterium-mediated transformation of sorghum (Sorghum bicolor (L.) Moench) using an improved in vitro regeneration system. Plant Cell Tiss Org Cult 91:155–164
Nishimura A (2020) Agrobacterium transformation in the rice genome. In: Vaschetto L (ed) Cereal Genomics. Springer, New York, NY, pp 207–216
Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang H, Wang X, Wicker T, Bharti AK, Chapman J, Feltus FA, Gowik U, Grigoriev IV, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang L, Carpita NC, Freeling M, Gingle AR, Hash CT, Keller B, Klein P, Kresovich S, McCann MC, Ming R, Peterson DG, Mehboob ur R, Ware D, Westhoff P, Mayer KFX, Messing J, Rokhsar DS, (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Shahbandeh M (2021) Worldwide production of grain in 2020/21, by type. Statista. https://www.statista.com/statistics/263977/world-grain-production-by-type/. Cited 18 October 2021
Songstad DD, Petolino JF, Voytas DF, Reichert NA (2017) Genome editing of plants. Crit Rev Plant Sci 36:1–23
Visarada K, Kishore NS (2015) Advances in genetic transformation. In: Madhusudhana R, Rajendrakumar P, Patil JV (eds) Sorghum molecular breeding. Springer, New Delhi, India, pp 199–215
Williams S, Gray S, Laidlaw H, Godwin I (2004) Particle inflow gun-mediated transformation of Sorghum bicolor. In: Curtis IE (ed) Transgenic crops of the world. Springer, Dordecht, pp 89–102
Xin Z, Jiao Y, Chopra R, Gladman N, Burow G, Hayes C, Chen J, Emendack Y, Ware D, Burke J (2019) Pedigreed mutant library—a unique resource for sorghum improvement and genomics. In: Ciampitti IA, Prasad PV (eds) Sorghum: State of the Art and Future Perspectives. American Society of Agronomy, Madison, pp 73–96
Xin Z, Wang ML, Burow G, Burke J (2009) An induced sorghum mutant population suitable for bioenergy research. BioEnerg Res 2:10–16
Xiong Y, Zhang P, Warner R, Fang Z (2019) Sorghum grain: from genotype, nutrition, and phenolic profile to its health benefits and food applications. Compr Rev Food Science Food Safe 18:2025–2046
Yang M, Baral NR, Simmons BA, Mortimer JC, Shih PM, Scown CD (2020) Accumulation of high-value bioproducts in planta can improve the economics of advanced biofuels. Proc Natl Acad Sci USA 117:8639–8648
Zhao Z, Tomes D (2003) Sorghum transformation. Springer, Genetic transformation of plants, pp 91–107
Zhao Z-y, Cai T, Tagliani L, Miller M, Wang N, Pang H, Rudert M, Schroeder S, Hondred D, Seltzer J, Pierce D (2000) Agrobacterium-mediated sorghum transformation. Plant Mol Biol 44:789–798
Acknowledgements
This work was partially supported by the National Science Foundation Plant Genome Research Program (NSF PGRP) Grant #1444478 and by Department of Energy BER Grant #DE-SCOO18277 to APK. Special thanks to Geoff Williams, The Brown University Leduc Imaging Center, for assistance with the scanning electron microscopy.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nelson-Vasilchik, K., Hague, J.P., Tilelli, M. et al. Rapid transformation and plant regeneration of sorghum (Sorghum bicolor L.) mediated by altruistic Baby boom and Wuschel2. In Vitro Cell.Dev.Biol.-Plant 58, 331–342 (2022). https://doi.org/10.1007/s11627-021-10243-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11627-021-10243-8