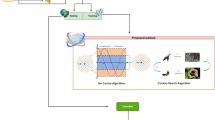
Abstract
Identifying a small subset of informative genes from a gene expression dataset is an important process for sample classification in the fields of bioinformatics and machine learning. In this process, there are two objectives: first, to minimize the number of selected genes, and second, to maximize the classification accuracy of the used classifier. In this paper, a hybrid machine learning framework based on a nature-inspired cuckoo search (CS) algorithm has been proposed to resolve this problem. The proposed framework is obtained by incorporating the cuckoo search (CS) algorithm with an artificial bee colony (ABC) in the exploitation and exploration of the genetic algorithm (GA). These strategies are used to maintain an appropriate balance between the exploitation and exploration phases of the ABC and GA algorithms in the search process. In preprocessing, the independent component analysis (ICA) method extracts the important genes from the dataset. Then, the proposed gene selection algorithms along with the Naive Bayes (NB) classifier and leave-one-out cross-validation (LOOCV) have been applied to find a small set of informative genes that maximize the classification accuracy. To conduct a comprehensive performance study, proposed algorithms have been applied on six benchmark datasets of gene expression. The experimental comparison shows that the proposed framework (ICA and CS-based hybrid algorithm with NB classifier) performs a deeper search in the iterative process, which can avoid premature convergence and produce better results compared to the previously published feature selection algorithm for the NB classifier.
Graphical abstract









Similar content being viewed by others
References
Ratan ZA et al (2018) CRISPR-Cas9: a promising genetic engineering approach in cancer research. Ther Adv Med Oncol 10:1758834018755089
Hartshorn CM et al (2018) Nanotechnology strategies to advance outcomes in clinical cancer care. ACS Nano 12(1):24–43
Halder A, Kumar A (2019) Active learning using rough fuzzy classifier for cancer prediction from microarray gene expression data. J Biomed Inform 92:103136
Rana HK et al (2020) Machine learning and bioinformatics models to identify pathways that mediate influences of welding fumes on cancer progression. Sci Rep 10(1):1–15
Shilo S, Rossman H, Segal E (2020) Axes of a revolution: challenges and promises of big data in healthcare. Nat Med 26(1):29–38
Cammarota G et al (2020) Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nat Rev Gastroenterol Hepatol 17(10):635–648
Qaraad M et al (2021) A hybrid feature selection optimization model for high dimension data classification. IEEE Access 9:42884–42895
Gumaei A et al (2021) Feature selection with ensemble learning for prostate cancer diagnosis from microarray gene expression. Health Inform J 27(1):1460458221989402
Lee J, Choi IY, Jun C-H (2021) An efficient multivariate feature ranking method for gene selection in high-dimensional microarray data. Expert Syst Appl 166:113971
Jain I, Jain VK, Jain R (2018) Correlation feature selection based improved-binary particle swarm optimization for gene selection and cancer classification. Appl Soft Comput 62:203–215
Wang H, Jing X, Niu B (2017) A discrete bacterial algorithm for feature selection in classification of microarray gene expression cancer data. Knowl-Based Syst 126:8–19
Aziz R, Verma C, Srivastava N (2016) A fuzzy based feature selection from independent component subspace for machine learning classification of microarray data. Genomics Data 8:4–15
Motwani A, Shukla PK, Pawar M (2021) Novel framework based on deep learning and cloud analytics for smart patient monitoring and recommendation (SPMR). J Ambient Intell Humaniz Comput 1:1–16
Lalwani P, Mishra MK, Chadha JS, Sethi P (2021) (pp 608-619). system: a machine learning approach. Computing 104(2):1–24
Aziz R, Verma CK, Srivastava N (2017) Dimension reduction methods for microarray data: a review. AIMS Bioeng 4(2):179–197
Valdez F, Castillo O, Peraza C (2020) Fuzzy logic in dynamic parameter adaptation of harmony search optimization for benchmark functions and fuzzy controllers. Int J Fuzzy Syst 22:1198–1211
Olivas F et al (2019) Interval type-2 fuzzy logic for dynamic parameter adaptation in a modified gravitational search algorithm. Inf Sci 476:159–175
Sanchez D, Melin P, Castillo O (2020) Comparison of particle swarm optimization variants with fuzzy dynamic parameter adaptation for modular granular neural networks for human recognition. J Intell Fuzzy Syst 38(3):3229–3252
Castillo O et al (2019) Comparative study in fuzzy controller optimization using bee colony, differential evolution, and harmony search algorithms. Algorithms 12(1):9
Lodh A, Saxena U, khan A, Motwani A, Shakkeera L, Sharmasth VY (2020) Prototype for integration of face mask detection and person identification model–COVID-19. In 2020 4th International Conference on Electronics, Communication and Aerospace Technology, IEEE
Castillo O, Melin P (2020) Forecasting of COVID-19 time series for countries in the world based on a hybrid approach combining the fractal dimension and fuzzy logic. Chaos, Solitons Fractals 140:110242
Sanchez MA, Castillo O, Castro JR (2015) Information granule formation via the concept of uncertainty-based information with interval type-2 fuzzy sets representation and Takagi–Sugeno–Kang consequents optimized with Cuckoo search. Appl Soft Comput 27:602–609
Khan ZA et al (2019) Hybrid meta-heuristic optimization based home energy management system in smart grid. J Ambient Intell Humaniz Comput 10(12):4837–4853
Singh RK, Sivabalakrishnan M (2015) Feature selection of gene expression data for cancer classification: a review. Procedia Comput Sci 50:52–57
Mafarja M et al (2020) Efficient hybrid nature-inspired binary optimizers for feature selection. Cogn Comput 12(1):150–175
Venkatesh B, Anuradha J (2019) A review of feature selection and its methods. Cybern Inform Technol 19(1):3–26
Sowmiya C, Sumitra P (2020) A hybrid approach for mortality prediction for heart patients using ACO-HKNN. J Ambient Intell Humaniz Comput 5(2021):1–8
Peng W et al (2020) Interval type-2 fuzzy logic based transmission power allocation strategy for lifetime maximization of WSNs. Eng Appl Artif Intell 87:103269
Ochoa P, Castillo O, Soria J (2020) Optimization of fuzzy controller design using a differential evolution algorithm with dynamic parameter adaptation based on type-1 and interval type-2 fuzzy systems. Soft Comput 24(1):193–214
Semwal VB, Gaud N, Lalwani P, Bijalwan V, Alok Ak (2021) Pattern identification of different human joints for different human walking styles using inertial measurement unit (IMU) sensor. Artif Intell Rev 55(2):1–21
Castillo O, Hidalgo D, Cervantes L, Melin P, Soto RM (2020) Fuzzy parameter adaptation in genetic algorithms for the optimization of fuzzy integrators in modular neural networks for multimodal biometry. Comput Sistemas 24(3):1093–105.
Tarek S, Abd Elwahab R, Shoman M (2017) Gene expression based cancer classification. Egypt Inform J 18(3):151–159
Gao L, Ye M, Wu C (2017) Cancer classification based on support vector machine optimized by particle swarm optimization and artificial bee colony. Molecules 22(12):2086
Chandrashekar G, Sahin F (2014) A survey on feature selection methods. Comput Electr Eng 40(1):16–28
Alshamlan HM, Badr GH, Alohali YA (2015) Genetic bee colony (GBC) algorithm: a new gene selection method for microarray cancer classification. Comput Biol Chem 56:49–60
Mahapatra B, Nayyar A (2019) Swarm intelligence and evolutionary algorithms for cancer diagnosis. In: Swarm Intelligence and Evolutionary Algorithms in Healthcare and Drug Development, vol 19
Sampathkumar A et al (2020) An efficient hybrid methodology for detection of cancer-causing gene using CSC for micro array data. J Ambient Intell Humaniz Comput 11(11):4743–4751
Gu S, Cheng R, Jin Y (2018) Feature selection for high-dimensional classification using a competitive swarm optimizer. Soft Comput 22(3):811–822
Hyvärinen A, Oja E (2000) Independent component analysis: algorithms and applications. Neural Netw 13(4-5):411–430
Musheer RA, Verma CK, Srivastava N (2019) Novel machine learning approach for classification of high-dimensional microarray data. Soft Comput 23(24):13409–13421
Kong W et al (2008) A review of independent component analysis application to microarray gene expression data. Biotechniques 45(5):501–520
Arowolo MO et al (2020) A hybrid heuristic dimensionality reduction methods for classifying malaria vector gene expression data. IEEE Access 8:182422–182430
Fan L, Poh K-L, Zhou P (2009) A sequential feature extraction approach for naïve bayes classification of microarray data. Expert Syst Appl 36(6):9919–9923
Mollaee M, Moattar MH (2016) A novel feature extraction approach based on ensemble feature selection and modified discriminant independent component analysis for microarray data classification. Biocybernetics Biomed Eng 36(3):521–529
Mahdavi K, Labarta J, Gimenez J (2019) Unsupervised feature selection for noisy data. In International Conference on Advanced Data Mining and Applications (pp. 79-94). Springer, Cham.
Aziz R et al (2017) Artificial neural network classification of microarray data using new hybrid gene selection method. Int J Data Min Bioinform 17(1):42–65
Aziz R, Verma CK, Srivastava N (2017) A novel approach for dimension reduction of microarray. Comput Biol Chem 71:161–169
Aziz R, Srivastava N, Verma CK (2015) T-independent component analysis for svm classification of dna-microarray data. Int J Bioinform Res, 3(2015):0975–3087
Pandey AC, Rajpoot DS, Saraswat M (2020) Feature selection method based on hybrid data transformation and binary binomial cuckoo search. J Ambient Intell Humaniz Comput 11(2):719–738
Cui Z et al (2019) A hybrid many-objective cuckoo search algorithm. Soft Comput 23(21):10681–10697
Peng H et al (2021) Multi-strategy serial cuckoo search algorithm for global optimization. Knowl-Based Syst 214:106729
Pandey AC, Rajpoot DS (2019) Spam review detection using spiral cuckoo search clustering method. Evol Intel 12(2):147–164
Cristin R, Kumar BS, Priya C, Karthick K (2020) Deep neural network based rider-cuckoo search algorithm for plant disease detection. Artif Intell Rev 53(7):1–26
Song P-C, Pan J-S, Chu S-C (2020) A parallel compact cuckoo search algorithm for three-dimensional path planning. Appl Soft Comput 94:106443
Zhang Z, Ding S, Jia W (2019) A hybrid optimization algorithm based on cuckoo search and differential evolution for solving constrained engineering problems. Eng Appl Artif Intell 85:254–268
Coleto-Alcudia V, Vega-Rodríguez MA (2020) Artificial bee colony algorithm based on dominance (ABCD) for a hybrid gene selection method. Knowl-Based Syst 205:106323
Wang X-h et al (2020) Multi-objective feature selection based on artificial bee colony: an acceleration approach with variable sample size. Appl Soft Comput 88:106041
Garro BA, Rodríguez K, Vázquez RA (2016) Classification of DNA microarrays using artificial neural networks and ABC algorithm. Appl Soft Comput 38:548–560
Hsu C-C, Chen M-C, Chen L-S (2010) Integrating independent component analysis and support vector machine for multivariate process monitoring. Comput Ind Eng 59(1):145–156
Karaboga D (2005) An idea based on honey bee swarm for numerical optimization. Citeseer
Alshamlan H, Badr G, Alohali Y (2015) mRMR-ABC: a hybrid gene selection algorithm for cancer classification using microarray gene expression profiling, (Article ID 604910), Biomed Res Int, volume (2015):1-16,
Abdel-Basset M, Hessin A-N, Abdel-Fatah L (2018) A comprehensive study of cuckoo-inspired algorithms. Neural Comput & Applic 29(2):345–361
Friedman N, Geiger D, Goldszmidt M (1997) Bayesian network classifiers. Mach Learn 29(2):131–163
Hall M (2006) A decision tree-based attribute weighting filter for naive Bayes. In International conference on innovative techniques and applications of artificial intelligence (pp. 59-70). Springer, London.
Chen J et al (2009) Feature selection for text classification with Naïve Bayes. Expert Syst Appl 36(3):5432–5435
Sandberg R et al (2001) Capturing whole-genome characteristics in short sequences using a naive Bayesian classifier. Genome Res 11(8):1404–1409
Fan L, Poh K-L, Zhou P (2010) Partition-conditional ICA for Bayesian classification of microarray data. Expert Syst Appl 37(12):8188–8192
De Campos LM, Cano A, Castellano JG, Moral S (2011) Bayesian networks classifiers for gene-expression data. In 2011 11th International Conference on Intelligent Systems Design and Applications, pp. 1200-1206. IEEE
Alon U et al (1999) Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc Natl Acad Sci 96(12):6745–6750
Golub TR et al (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286(5439):531–537
Singh D et al (2002) Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 1(2):203–209
Nutt CL et al (2003) Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res 63(7):1602–1607
Gordon GJ et al (2002) Translation of microarray data into clinically relevant cancer diagnostic tests using gene expression ratios in lung cancer and mesothelioma. Cancer Res 62(17):4963–4967
Armstrong SA et al (2002) MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet 30(1):41–47
Aziz R, Verma C, Srivastava N (2015) A weighted-SNR feature selection from independent component subspace for nb classification of microarray data. Int J Adv Biotechnol Res 6:245–255
Xi M et al (2016) Cancer feature selection and classification using a binary quantum-behaved particle swarm optimization and support vector machine. Comput Math Methods Med 2016
Akay B, Karaboga D, (2009) Parameter tuning for the artificial bee colony algorithm. In International conference on computational collective intelligence. Springer, Berlin, Heidelberg pp 608–619
Varghese MP, Amudha A (2018) Artificial Bee Colony and Cuckoo Search Algorithm for Cost Estimation with Wind Power Energy. Int J Simul Syst Sci Technol 19(6). https://doi.org/10.5013/IJSSST.a.19.06.18
Raczko E, Zagajewski B (2017) Comparison of support vector machine, random forest and neural network classifiers for tree species classification on airborne hyperspectral APEX images. Eur J Remote Sens 50(1):144–154
Huang M-W et al (2017) SVM and SVM ensembles in breast cancer prediction. PLoS One 12(1):e0161501
Nahar J, Ali S, Chen Y-PP (2007) Microarray data classification using automatic SVM kernel selection. DNA Cell Biol 26(10):707–712
Aziz R, Verma CK, Srivastava N (2018) Artificial neural network classification of high dimensional data with novel optimization approach of dimension reduction. Ann Data Sci 5(4):615–635
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Aziz, R.M. Nature-inspired metaheuristics model for gene selection and classification of biomedical microarray data. Med Biol Eng Comput 60, 1627–1646 (2022). https://doi.org/10.1007/s11517-022-02555-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-022-02555-7