Abstract
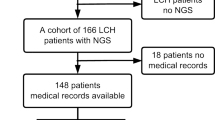
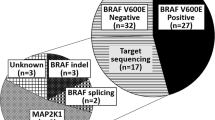
ARAF mutations have been identified in a limited subset of patients with Langerhans cell histiocytosis (LCH), a rare disorder characterized by abnormal proliferation of Langerhans cells. LCH is primarily instigated by mutations in the mitogen-activated protein kinase (MAPK) signaling pathway, with BRAFV600E and MAP2K1 mutations constituting most cases. ARAF mutations in LCH highlight the heterogeneity of the disease and provide insights into its underlying molecular mechanisms. However, the occurrence of ARAF-positive LCH cases is extremely rare, with only two reported globally. Although they may be linked to a more aggressive form of LCH and a more severe clinical progression, the clinical significance and functional consequences of these mutations remain uncertain. We performed next-generation sequencing (NGS) to explore driver mutations in 148 pediatric LCH patients and recognized a series of mutations, including an identical novel somatic ARAF mutation, c.1046_1051delAGGCTT (p.Q349_F351delinsL), in four pediatric LCH patients. It was considered an ARAF hotspot mutation. All reported ARAF-positive patients worldwide exhibited characteristic pathological features of LCH, albeit with involvement across multiple systems. In vitro functional studies showed that this mutation could trigger the MAPKinase pathway and phosphorylate its downstream effectors MEK1/2 and ERK1/2 (relatively weaker than BRAFV600E). Over-activation of mutant A-Raf kinase could be inhibited by the BRAF inhibitor vemurafenib. LCH is uncommon, and ARAF mutation is even rarer. In our study, we have identified a novel hotspot somatic ARAF mutation, which has been verified through functional analysis to be an activating mutation. LCH patients with ARAF mutation typically have an unfavorable prognosis due to limited treatment experiences, although they do not exhibit a high relapse rate. To aid in the development of personalized treatment approaches and prognostic markers for LCH patients, it is recommended to conduct typical pathological and immunohistochemical examinations, as well as genetic tests utilizing a targeted gene panel or whole exome sequencing (WES), for LCH diagnosis, thereby promoting the use of inhibitor treatment strategies.




Similar content being viewed by others
Data availability statement
The datasets used to support this paper's results are available in the article and more data will be available upon request. Additional sequencing data files have been deposited in NCBI GenBank under accession number PRJNA875244.
Abbreviations
- DCs:
-
Dendritic cells
- ECD:
-
Erdheim–Chester disease
- ERK:
-
Extracellular signal-regulated kinase
- FFPE:
-
Formalin-fixed paraffin-embedded
- IHC:
-
Histopathology and immunohistochemistry
- LCH:
-
Langerhans cell histiocytosis
- MAPKinase:
-
Mitogen-activated protein kinase
- MEK:
-
Mitogen-activated protein kinase kinase
- MS:
-
Multiple-system involvement
- MS-OR− :
-
Multiple-system without risk-organ involvement
- MS-OR+ :
-
Multiple-system with risk-organ involvement
- NGS:
-
Next-generation sequencing
- SS:
-
Single system involvement
- WES:
-
Whole exome sequencing
References
Allen CE, Merad M, McClain KL. Langerhans-Cell Histiocytosis. N Engl J Med. 2018;379(9):856–68.
Rodriguez-Galindo C, Allen CE. Langerhans cell histiocytosis. Blood. 2020;135(16):1319–31.
Degar BA, Rollins BJ. Langerhans cell histiocytosis: malignancy or inflammatory disorder doing a great job of imitating one? Dis Model Mech. 2009;2(9–10):436–9.
Li Z, Yanqiu L, Yan W, et al. Two case report studies of Langerhans cell histiocytosis with an analysis of 918 patients of Langerhans cell histiocytosis in literatures published in China. Int J Dermatol. 2010;49(10):1169–74.
Harmon CM, Brown N. Langerhans cell histiocytosis: a clinicopathologic review and molecular pathogenetic update. Arch Pathol Lab Med. 2015;139(10):1211–4.
Gulati N, Allen CE. Langerhans cell histiocytosis: Version 2021. Hematol Oncol. 2021;39(Suppl 1):15–23.
Berres ML, Merad M, Allen CE. Progress in understanding the pathogenesis of Langerhans cell histiocytosis: back to Histiocytosis X? Br J Haematol. 2015;169(1):3–13.
Ballester LY, Cantu MD, Lim KPH, et al. The use of BRAF V600E mutation-specific immunohistochemistry in pediatric Langerhans cell histiocytosis. Hematol Oncol. 2018;36(1):307–15.
Heritier S, Emile JF, Barkaoui MA, et al. BRAF mutation correlates with high-risk langerhans cell histiocytosis and increased resistance to first-line therapy. J Clin Oncol. 2016;34(25):3023–30.
Kobayashi M, Ando S, Kawamata T, et al. Clinical features and outcomes of adult Langerhans cell histiocytosis: a single-center experience. Int J Hematol. 2020;112(2):185–92.
Liu X, Zhang Y, Zhou CX. High prevalence of BRAF V600E mutations in langerhans cell histiocytosis of head and neck in chinese patients. Int J Surg Pathol. 2019;27(8):836–43.
Sasaki Y, Guo Y, Arakawa F, et al. Analysis of the BRAFV600E mutation in 19 cases of Langerhans cell histiocytosis in Japan. Hematol Oncol. 2017;35(3):329–34.
Feng S, Han L, Yue M, et al. Frequency detection of BRAF V600E mutation in a cohort of pediatric langerhans cell histiocytosis patients by next-generation sequencing. Orphanet J Rare Dis. 2021;16(1):272.
Krooks J, Minkov M, Weatherall AG. Langerhans cell histiocytosis in children: History, classification, pathobiology, clinical manifestations, and prognosis. J Am Acad Dermatol. 2018;78(6):1035–44.
Haupt R, Minkov M, Astigarraga I, et al. Langerhans cell histiocytosis (LCH): guidelines for diagnosis, clinical work-up, and treatment for patients till the age of 18 years. Pediatr Blood Cancer. 2013;60(2):175–84.
Minkov M, Grois N, Heitger A, et al. Treatment of multisystem Langerhans cell histiocytosis. Results of the DAL-HX 83 and DAL-HX 90 studies. DAL-HX Study Group. Klin Padiatr. 2000;212(4):139–44.
Morimoto A, Ikushima S, Kinugawa N, et al. Improved outcome in the treatment of pediatric multifocal Langerhans cell histiocytosis: Results from the Japan Langerhans Cell Histiocytosis Study Group-96 protocol study. Cancer. 2006;107(3):613–9.
Gadner H, Minkov M, Grois N, et al. Therapy prolongation improves outcome in multisystem Langerhans cell histiocytosis. Blood. 2013;121(25):5006–14.
Sakamoto K, Morimoto A, Shioda Y, et al. Long-term complications in uniformly treated paediatric Langerhans histiocytosis patients disclosed by 12 years of follow-up of the JLSG-96/02 studies. Br J Haematol. 2021;192(3):615–20.
Morimoto A, Shioda Y, Imamura T, et al. Intensification of induction therapy and prolongation of maintenance therapy did not improve the outcome of pediatric Langerhans cell histiocytosis with single-system multifocal bone lesions: results of the Japan Langerhans Cell Histiocytosis Study Group-02 Protocol Study. Int J Hematol. 2018;108(2):192–8.
Sondka Z, Bamford S, Cole CG, et al. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer. 2018;18(11):696–705.
Steinhaus R, Proft S, Schuelke M, et al. MutationTaster2021. Nucleic Acids Res. 2021;49(W1):W446–51.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–81.
Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31(16):2745–7.
Ferlaino M, Rogers MF, Shihab HA, et al. An integrative approach to predicting the functional effects of small indels in non-coding regions of the human genome. BMC Bioinformatics. 2017;18(1):442.
Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–94.
Li S, van der Velde KJ, de Ridder D, et al. CAPICE: a computational method for Consequence-Agnostic Pathogenicity Interpretation of Clinical Exome variations. Genome Med. 2020;12(1):75.
Siepel A, Bejerano G, Pedersen JS, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–50.
Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20(1):110–21.
Li MM, Datto M, Duncavage EJ, et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19(1):4–23.
Berres ML, Lim KP, Peters T, et al. BRAF-V600E expression in precursor versus differentiated dendritic cells defines clinically distinct LCH risk groups. J Exp Med. 2014;211(4):669–83.
Satoh T, Smith A, Sarde A, et al. B-RAF mutant alleles associated with Langerhans cell histiocytosis, a granulomatous pediatric disease. PLoS ONE. 2012;7(4): e33891.
Badalian-Very G, Vergilio JA, Degar BA, Rodriguez-Galindo C, Rollins BJ. Recent advances in the understanding of Langerhans cell histiocytosis. Br J Haematol. 2012;156(2):163–72.
DiCaprio MR, Roberts TT. Diagnosis and management of Langerhans cell histiocytosis. J Am Acad Orthop Surg. 2014;22(10):643–52.
Gan X, Wang J, Wang C, et al. PRR5L degradation promotes mTORC2-mediated PKC-delta phosphorylation and cell migration downstream of Galpha12. Nat Cell Biol. 2012;14(7):686–96.
Consortium APG. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017;7(8):818–31.
Nelson DS, Quispel W, Badalian-Very G, et al. Somatic activating ARAF mutations in Langerhans cell histiocytosis. Blood. 2014;123(20):3152–5.
Chakraborty R, Hampton OA, Shen X, et al. Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood. 2014;124(19):3007–15.
McGinnis LM, Nybakken G, Ma L, Arber DA. Frequency of MAP2K1, TP53, and U2AF1 mutations in BRAF-mutated Langerhans Cell histiocytosis: further characterizing the genomic landscape of LCH. Am J Surg Pathol. 2018;42(7):885–90.
Diamond EL, Durham BH, Haroche J, et al. Diverse and targetable kinase alterations drive histiocytic neoplasms. Cancer Discov. 2016;6(2):154–65.
Rebocho AP, Marais R. ARAF acts as a scaffold to stabilize BRAF:CRAF heterodimers. Oncogene. 2013;32(26):3207–12.
Li D, March ME, Gutierrez-Uzquiza A, et al. ARAF recurrent mutation causes central conducting lymphatic anomaly treatable with a MEK inhibitor. Nat Med. 2019;25(7):1116–22.
Brown NA, Furtado LV, Betz BL, et al. High prevalence of somatic MAP2K1 mutations in BRAF V600E-negative Langerhans cell histiocytosis. Blood. 2014;124(10):1655–8.
Chakraborty R, Burke TM, Hampton OA, et al. Alternative genetic mechanisms of BRAF activation in Langerhans cell histiocytosis. Blood. 2016;128(21):2533–7.
Heritier S, Helias-Rodzewicz Z, Chakraborty R, et al. New somatic BRAF splicing mutation in Langerhans cell histiocytosis. Mol Cancer. 2017;16(1):115.
Nelson DS, van Halteren A, Quispel WT, et al. MAP2K1 and MAP3K1 mutations in Langerhans cell histiocytosis. Genes Chromosomes Cancer. 2015;54(6):361–8.
Baljuls A, Mueller T, Drexler HC, Hekman M, Rapp UR. Unique N-region determines low basal activity and limited inducibility of A-RAF kinase: the role of N-region in the evolutionary divergence of RAF kinase function in vertebrates. J Biol Chem. 2007;282(36):26575–90.
Hogstad B, Berres ML, Chakraborty R, et al. RAF/MEK/extracellular signal-related kinase pathway suppresses dendritic cell migration and traps dendritic cells in Langerhans cell histiocytosis lesions. J Exp Med. 2018;215(1):319–36.
Morimoto A, Shioda Y, Imamura T, et al. Intensified and prolonged therapy comprising cytarabine, vincristine and prednisolone improves outcome in patients with multisystem Langerhans cell histiocytosis: results of the Japan Langerhans Cell Histiocytosis Study Group-02 Protocol Study. Int J Hematol. 2016;104(1):99–109.
Diamond EL, Subbiah V, Lockhart AC, et al. Vemurafenib for BRAF V600-Mutant Erdheim-Chester disease and Langerhans cell histiocytosis: analysis of data from the histology-independent, Phase 2. Open-label VE-BASKET Study JAMA Oncol. 2018;4(3):384–8.
Diamond EL, Durham BH, Ulaner GA, et al. Efficacy of MEK inhibition in patients with histiocytic neoplasms. Nature. 2019;567(7749):521–4.
Eckstein OS, Visser J, Rodriguez-Galindo C, Allen CE. Clinical responses and persistent BRAF V600E(+) blood cells in children with LCH treated with MAPK pathway inhibition. Blood. 2019;133(15):1691–4.
Donadieu J, Larabi IA, Tardieu M, et al. Vemurafenib for refractory multisystem Langerhans cell Histiocytosis in children: an international observational study. J Clin Oncol. 2019;37(31):2857–65.
Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med. 2015;373(8):726–36.
Acknowledgements
We are grateful to patients and families for their cooperation.
Funding
The present study was supported by the Project from Wujieping Medical Foundation of China (Grant No. 20.6750.2021-04-37).
Author information
Authors and Affiliations
Author notes
Rong Liu and Yibing Guo are Co-first authors.
Contributions
RL designed research, collected and analyzed clinical data. YG designed research, analyzed data and wrote the manuscript. LH analyzed data and performed laboratory work. SF and JC contributed to the care and management of patients. SF also collected clinical data. YS is the liaison between patients and researchers. ZC and XC conceived and supervised research. All authors have approved the submitted manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors Y.G., L.H., Y.S. and Z.C. are employed by GrandOmics (Beijing, China). All authors declared no competing financial interests.
Ethics approval and consent to participate
This study was performed according to the Declaration of Helsinki, and the collection of tissues or blood samples from patients was obtained with patient-informed consent (from their guardians) under approval by the research ethics committee of the Children’s Hospital of Capital Institute of Pediatrics (Identifier: SHERLLM2020005). All patients provided written informed consent for publication of their cases.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, R., Guo, Y., Han, L. et al. Somatic ARAF mutations in pediatric Langerhans cell histiocytosis: clinicopathologic, genetic and functional profiling. Clin Exp Med 23, 5269–5279 (2023). https://doi.org/10.1007/s10238-023-01134-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10238-023-01134-w