Abstract
Background
Recent studies have uncovered that the aberrant expression of LINC00665 contributes to the malignant pathological process of various cancers and is closely related to the unfavorable prognosis of patients with cancer. However, a systematic analysis of the prognostic and clinicopathologic values of LINC00665 in cancers has not been conducted.
Objective
We aim to clarify the association of LINC00665 expression with patient survival and clinicopathologic phenotypes in cancers.
Methods
An electronic search of PubMed, Embase and Web of Science was performed to select eligible literature. Pooled hazard ratio (HR) and odds ratio (OR) were calculated to assess the clinical importance of LINC00665. The fixed-effects model was used to analyze the combined HR values and 95% CI when the studies had no significant heterogeneity (P > 0.1 for the Chi-square test or I2 < 50%). Begg’s test and sensitivity analysis were also conducted. This study was registered in The International Prospective Register of Systematic Reviews (PROSPERO registration number: CRD42021290123).
Results
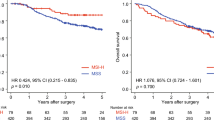
A total of 710 patients from 10 eligible studies were enrolled in this meta-analysis, which was based on China population. The pooled results of this analysis revealed that high-level expression of LINC00665 was notably correlated with poor overall survival (HR = 2.08, 95% CI = 1.57–2.75) and recurrence-free survival (HR = 2.49, 95% CI = 1.63–3.80) in human cancers. Elevated LINC00665 expression was also correlated with more advanced clinical stage, earlier lymph node metastasis, lower tumor differentiation, earlier distant metastasis and larger tumor size.
Conclusion
LINC00665 expression was critically related to the cancer prognosis, which has important prognostic implications for clinical prediction.






Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32.
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33.
Chi Y, Wang D, Wang J, Yu W, Yang J. Long non-coding RNA in the pathogenesis of cancers. Cells. 2019;8(9):1015.
Mansoori B, Mohammadi A, Davudian S, Shirjang S, Baradaran B. The different mechanisms of cancer drug resistance: a brief review. Adv Pharm Bull. 2017;7(3):339–48.
Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–8.
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–41.
Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–81.
Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300.
Chen L, Zhang YH, Lu G, Huang T, Cai YD. Analysis of cancer-related lncRNAs using gene ontology and KEGG pathways. Artif Intell Med. 2017;76:27–36.
Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–41.
Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29(3):212–26.
Peng F, Wang R, Zhang Y, et al. Differential expression analysis at the individual level reveals a lncRNA prognostic signature for lung adenocarcinoma. Mol Cancer. 2017;16(1):98.
Peng K, Liu R, Yu Y, et al. Identification and validation of cetuximab resistance associated long noncoding RNA biomarkers in metastatic colorectal cancer. Biomed Pharmacother. 2018;97:1138–46.
Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–63.
Smits G, Mungall AJ, Griffiths-Jones S, et al. Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nat Genet. 2008;40(8):971–6.
Zemel S, Bartolomei MS, Tilghman SM. Physical linkage of two mammalian imprinted genes, H19 and insulin-like growth factor 2. Nat Genet. 1992;2(1):61–5.
Pan Y, Zhang Y, Liu W, et al. LncRNA H19 overexpression induces bortezomib resistance in multiple myeloma by targeting MCL-1 via miR-29b-3p. Cell Death Dis. 2019;10(2):106.
Peperstraete E, Lecerf C, Collette J, et al. Enhancement of breast cancer cell aggressiveness by lncRNA H19 and its Mir-675 derivative: insight into shared and different actions. Cancers (Basel). 2020;12(7):1730.
Wang J, Xie S, Yang J, et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J Hematol Oncol. 2019;12(1):81.
Ren J, Ding L, Zhang D, et al. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8(14):3932–48.
Kamel MM, Matboli M, Sallam M, Montasser IF, Saad AS, El-Tawdi AHF. Investigation of long noncoding RNAs expression profile as potential serum biomarkers in patients with hepatocellular carcinoma. Transl Res. 2016;168:134–45.
Wen DY, Lin P, Pang YY, et al. Expression of the long intergenic non-protein coding RNA 665 (LINC00665) gene and the cell cycle in hepatocellular carcinoma using the cancer genome atlas, the gene expression omnibus, and quantitative real-time polymerase chain reaction. Med Sci Monit. 2018;24:2786–808.
Guo B, Wu S, Zhu X, et al. Micropeptide CIP2A-BP encoded by LINC00665 inhibits triple-negative breast cancer progression. EMBO J. 2020;39(1): e102190.
Wan H, Tian Y, Zhao J, Su X. LINC00665 targets miR-214-3p/MAPK1 axis to accelerate hepatocellular carcinoma growth and warburg effect. J Oncol. 2021;2021:9046798.
Yang D, Feng W, Zhuang Y, et al. Long non-coding RNA linc00665 inhibits CDKN1C expression by binding to EZH2 and affects cisplatin sensitivity of NSCLC cells. Mol Ther Nucleic Acids. 2021;23:1053–65.
Lv M, Mao Q, Li J, Qiao J, Chen X, Luo S. Knockdown of LINC00665 inhibits proliferation and invasion of breast cancer via competitive binding of miR-3619-5p and inhibition of catenin beta 1. Cell Mol Biol Lett. 2020;25:43.
Cai Y, Hao M, Chang Y, Liu Y. LINC00665 enhances tumorigenicity of endometrial carcinoma by interacting with high mobility group AT-hook 1. Cancer Cell Int. 2021;21(1):8.
Ruan X, Zheng J, Liu X, et al. lncRNA LINC00665 stabilized by TAF15 impeded the malignant biological behaviors of glioma cells via STAU1-mediated mRNA degradation. Mol Ther Nucleic Acids. 2020;20:823–40.
Moher D, Shamseer L, Clarke M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev. 2015;4(1):1.
Wells GA, Shea B, O'Connell D, et al. The Newcastle‐Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in metaanalyses. Ottawa Hospital Research Institute. http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed 30 November 2021.
Cong Z, Diao Y, Xu Y, et al. Long non-coding RNA linc00665 promotes lung adenocarcinoma progression and functions as ceRNA to regulate AKR1B10-ERK signaling by sponging miR-98. Cell Death Dis. 2019;10(2):84.
Xu D, Song Q, Liu Y, et al. LINC00665 promotes ovarian cancer progression through regulating the miRNA-34a-5p/E2F3 axis. J Cancer. 2021;12(6):1755–63.
Dai Y, Zhang Y, Hao M, Zhu R. LINC00665 functions as a competitive endogenous RNA to regulate AGTR1 expression by sponging miR-34a-5p in glioma. Oncol Rep. 2021;45(3):1202–12.
Ding J, Zhao J, Huan L, et al. Inflammation-induced long intergenic noncoding RNA (LINC00665) increases malignancy through activating the double-stranded RNA-activated protein kinase/nuclear factor kappa B pathway in hepatocellular carcinoma. Hepatology. 2020;72(5):1666–81.
Zhang X, Wu J. LINC00665 promotes cell proliferation, invasion, and metastasis by activating the TGF-β pathway in gastric cancer. Pathol Res Pract. 2021;224: 153492.
Qi L, Sun B, Yang B, Lu S. LINC00665 stimulates breast cancer progression via regulating miR-551b-5p. Cancer Manag Res. 2021;13:1113–21.
Wang L, Song X, Yu L, Liu B, Ma J, Yang W. LINC00665 facilitates the malignant processes of osteosarcoma by increasing the RAP1B expression via sponging miR-708 and miR-142-5p. Anal Cell Pathol (Amst). 2021;2021:5525711.
Xue P, Yan M, Wang K, Gu J, Zhong B, Tu C. Up-Regulation of LINC00665 facilitates the malignant progression of prostate cancer by epigenetically silencing KLF2 through EZH2 and LSD1. Front Oncol. 2021;11: 639060.
Lu M, Qin X, Zhou Y, et al. Long non-coding RNA LINC00665 promotes gemcitabine resistance of cholangiocarcinoma cells via regulating EMT and stemness properties through miR-424-5p/BCL9L axis. Cell Death Dis. 2021;12(1):72.
Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinform. 2016;14(1):42–54.
Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19(3):143–57.
Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90.
Yao X, Lan Z, Lai Q, Li A, Liu S, Wang X. LncRNA SNHG6 plays an oncogenic role in colorectal cancer and can be used as a prognostic biomarker for solid tumors. J Cell Physiol. 2020;235(10):7620–34.
Jiang YF, Zhang HY, Ke J, Shen H, Ou HB, Liu Y. Overexpression of LncRNA GHET1 predicts an unfavourable survival and clinical parameters of patients in various cancers. J Cell Mol Med. 2019;23(8):4891–9.
Mehrad-Majd H, Akhtari J, Haerian MS, Ravanshad Y. Clinicopathological and prognostic value of lncRNA PANDAR expression in solid tumors: evidence from a systematic review and meta-analysis. J Cell Physiol. 2019;234(4):4206–16.
Xiao M, Feng Y, Liu C, Zhang Z. Prognostic values of long noncoding RNA PVT1 in various carcinomas: an updated systematic review and meta-analysis. Cell Prolif. 2018;51(6): e12519.
Liu X, Lu X, Zhen F, et al. LINC00665 induces acquired resistance to gefitinib through recruiting EZH2 and activating PI3K/AKT pathway in NSCLC. Mol Ther Nucleic Acids. 2019;16:155–61.
Wang H, Wang L, Zhang S, Xu Z, Zhang G. Downregulation of LINC00665 confers decreased cell proliferation and invasion via the miR-138-5p/E2F3 signaling pathway in NSCLC. Biomed Pharmacother. 2020;127: 110214.
Wang A, Zhang T, Wei W, et al. The long noncoding RNA LINC00665 FACILITATES c-Myc transcriptional activity via the miR-195-5p MYCBP axis to promote progression of lung adenocarcinoma. Front Oncol. 2021;11: 666551.
Ji W, Diao YL, Qiu YR, Ge J, Cao XC, Yu Y. LINC00665 promotes breast cancer progression through regulation of the miR-379–5p/LIN28B axis. Cell Death Dis. 2020;11(1):1–11.
Nan S, Zhang S, Jin R, Wang J. LINC00665 up-regulates SIN3A expression to modulate the progression of colorectal cancer via sponging miR-138-5p. Cancer Cell Int. 2022;22(1):51.
Zhang DW, Gu GQ, Chen XY, Zha GC, Yuan Z, Wu Y. LINC00665 facilitates the progression of osteosarcoma via sponging miR-3619-5p. Eur Rev Med Pharmacol Sci. 2020;24(19):9852–9.
Zhong WZ, Wang Q, Mao WM, et al. Gefitinib versus vinorelbine plus cisplatin as adjuvant treatment for stage II-IIIA (N1–N2) EGFR-mutant NSCLC: final overall survival analysis of CTONG1104 phase III trial. J Clin Oncol. 2021;39(7):713–22.
Dai H, Sheng X, Sha R, et al. Linc00665 can predict the response to cisplatin-paclitaxel neoadjuvant chemotherapy for breast cancer patients. Front Oncol. 2021;11: 604319.
Eke I, Bylicky MA, Sandfort V, et al. The lncRNAs LINC00261 and LINC00665 are upregulated in long-term prostate cancer adaptation after radiotherapy. Mol Ther Nucleic Acids. 2021;24:175–87.
Funding
This study was supported by the National Key Research and Development Program of China (2018YFC2002000).
Author information
Authors and Affiliations
Contributions
ZJ designed the study and wrote the manuscript; ZJ and YJM searched literatures and extracted the data; Two reviewers (MMW, DC) assessed study quality, and disputes are regulated by YSX. ZJ, YSX and XJ performed the data analyses; ZFX helped perform the analysis with constructive discussions. All authors did the final approval of the version to be published and agreed to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Conflict of interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors. Therefore, no ethical approval is required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jin, Z., Meng, YJ., Xu, YS. et al. Prognostic and clinicopathological values of LINC00665 in cancers: a systematic review and China population-based meta-analysis. Clin Exp Med 23, 1475–1487 (2023). https://doi.org/10.1007/s10238-022-00912-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10238-022-00912-2