Abstract
Background
CENPK is a novel oncogene which is aberrantly expression in some malignant tumors. However, the role and mechanisms of CENPK in gastric cancer have not been explored.
Methods
In this study, we use RT-PCR and IHC to study CENPK expression in gastric cancer cells and tissues. In addition, we constructed the two kinds of CENPK siRNA lentivirus to knock down CENPK. Then, we use High content living cell imaging System, Cell Counting Kit-8, colony formation, wound healing and Transwell assays to demonstrate the function of CENPK on gastric cancer cells AGS and MKN45. Meanwhile, we use flow cytometry assay to study CENPK function on gastric cancer cell apoptosis and cell cycle arrest. Subcutaneous tumorigenesis in nude mice was also performed to confirm CENPK function on gastric cancer. Finally, we use Co-IP, LC-MS and function rescue assay to study the downstream interaction molecular of CENPK.
Results
We demonstrated that CENPK expression were up-regulated in GC cell lines. Poor differentiation and III-IV stage had more percentages of high CENPK expression. Knocking down CENPK could significantly suppress GC cells proliferation, migration and invasion, and induce GC cells apoptosis and G1/S phase transition arrest. Subcutaneous tumorigenesis confirmed the tumor-promoting effects of CENPK in vivo. Remarkably, we found for the first time that XRCC5 might be interacted with CENPK through Co-IP, LC-MS and rescue study.
Conclusion
CENPK promotes GC cell proliferation and migration via interacting with XRCC5 and may be a novel prognostic factor or therapeutic target for CENPK.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Gastric cancer (GC) ranks fifth in global cancer incidence and third in cancer-related mortality [1]. Its incidence rate and mortality rate are the first in digestive tract cancer. However, the detection rate of early gastric cancer is low. Most gastric cancer is in the middle and advanced stage when founded, and some patients lose the opportunity of surgical treatment [2]. Therefore, chemotherapy-based comprehensive treatment is the main treatment for this kind of patients. A comprehensive chemotherapy based on 5-FU is suggested. However, there are many deficiencies in the clinical application of 5-FU, such as drug resistance, high toxicity and side effects, so the therapeutic effect and survival time of patients with gastric cancer are restricted. At present, the effective targeted drugs for gastric cancer is still deficient. Therefore, it is very important to further clarify the occurrence and development mechanism of gastric cancer and find new effective anti-gastric cancer-targeted therapy to improve the treatment strategy of gastric cancer.
TCGA (the Cancer Genome Atlas) database, also known as the cancer and tumor gene mapping project, integrates all human cancers through the application of genome analysis technology, especially large-scale genome sequencing (the short-term goal is to draw the genome variation map of 50 kinds of tumors including subtypes) and conduct systematic analysis, to find the small variation of all carcinogenic and tumor suppressor genes, understand the mechanism of cancer cell occurrence and development, obtain new diagnosis and treatment methods on this basis, and finally outline the whole new “cancer prevention strategy”. The project is jointly carried out by the National Cancer and Cancer Institute (NCI) and the National Human Genome Institute (NHGRI). In this study, we found that expression of CENPK (Centromeric protein K) gene was significantly up-regulated in gastric cancer in TCGA database.
CENPK is a member of the centromeric protein family, which is closely related to normal mitosis of cells [3], and participates in the formation of nucleoplasm in intercellular phase [3,4,5]. In recent years, studies have found that the overexpression of CENPK is related to the occurrence and development of tumors [6] and pulmonary fibrosis [7], such as melanoma [5], ovarian cancer [8], liver cancer [9, 10], lung cancer [11]. However, whether CENPK is closely related with the progression of gastric cancer is not clear, and how does CENPK play function in the tumor oncogenes is not reported until now.
To further investigate the effect of CENPK gene on the occurrence and development of gastric cancer, in this study, we designed and knocked down CENPK with two kinds of siRNA sequence to study its role in gastric cancer cell both in vitro and in vivo. Furthermore, we explore Co-IP (Co-Immunoprecipitation) and LC-MS (Liquid Chromatograph-Mass Spectrometry) to further identified the potential downstream proteins of CENPK.
The study will further clarify the role and clinical relevance of CENPK gene on the occurrence and development of gastric cancer, as well as clarify the molecular mechanism of CENPK, therefore provide a theoretical basis for CENPK gene as a potential therapeutic target for gastric cancer.
Methods
Bioinformatics analysis
The clinical information and RNAseqV2 data of 416 stomach adenocarcinoma (STAD) patients were downloaded from the TCGA database using the GDC Data Portal (https://gdc-portal.nci.nih.gov). There were 32 cases with RNAseq V2 paired sample data and pathological information (Supplementary Table1). TMM (trimmed mean of m-values) method was used for data standardization. We performed a quality control by observing BCV (biological coefficient of variation) and choose 26 cases with high stability for subsequence procession. The differently express genes (DEGs) between STAD and normal tissues with the cut-off criteria of P < 0.05 and log2 FC (fold-change)>1.0 were identified.
Patients and specimens
A total of 63 pairs of GC (gastric cancer) and paracancerous tissue samples of patients underwent initial surgical resection were obtained from January 2018 and December 2020 at Gansu Provincial Hospital, and were analyzed with Immunohistochemistry (IHC). All patients were diagnosed with gastric cancer by two independent pathologists. This study had been approved by the Ethics review committee of Gansu Provincial Hospital, and informed consents had been obtained from each patient.
Immunohistochemistry (IHC)
Paraffin sections were made, dehydrated with gradient alcohol, and then antigen was repaired by thermal repair method to eliminate the activity of endogenous peroxidase, then the tissue slides were blocked with goat serum and incubated with primary antibodies (CENPK, BIOSS, China; ki-67, CST, USA) overnight at 4 °C. The next day slides were treated with secondary antibodies (ZSGB-BIO, China, SP-9001) and colored by DAB peroxidase substrate kit for IHC (ZSGB-BIO, China, ZLI 9018). The slides were observed and photographed under Ortho fluorescence microscope (Olympus BX43).
Cell lines and cell culture
AGS, BGC-823, MGC80-3, SGC-7901, and MKN45 gastric cancer lines were purchased from the Chinese Academy of Science Cell Bank (Shanghai, China). All the cell lines were cultured with Dulbecco's modified Eagle's medium (DMEM) (Corning, USA) supplemented with 10% fetal bovine serum (Ausbian, USA) and 100 μg/ml penicillin G and streptomycin (Sigma, Shanghai, China) at 37 °C in a humidified incubator containing 5% CO2. AGS and MKN45 cells were used in proliferation, migration and apoptosis assay, MKN45 cell lines were used in vivo tumorigenic assay, and AGS cells were used in CO-IP and LC-MS assay.
Lentivirus construction and infection
CENPK short hairpin RNAs (shCENPK1 and shCENKP2) and the scramble shRNA control were constructed and purchased from GeneChem Company (Shanghai, China). The target sequences of the shRNAs were as follows: shCENPK-1: 5′-AATGTGGAAAGATATGGAA-3′ and shCENPK-2: 5′-ACCTTACCTCATGCAATAT-3′. The scramble shRNA control sequences were as follows: TTCTCCGAACGTGTCACGT. RNAi lentivirus vector were constructed with GV lentivirus vector series containing GFP, pahelper 1.0 vector and Phelper 2.0 carrier (GeneChem Company, Shanghai, China) in 293T cells. AGS and MKN45 cells were infected with the shCENPK-1 and shCENKP-2 lentivirus using polybrene (5 μg/ml) and subsequently selected with puromycin (5 μg/ml) for 5 days to establish the stable CENPK-Knockdown cell lines. Cells transfected with scramble vector lentivirus were used as controls. After transfection, expression of CENPK in gastric cell lines were detected by qRT-PCR and western blot.
RNA extraction and qRT-PCR
Total RNA of different cell lines were extracted and purified using Trizol (Lifetechnologies, USA) and cDNA was synthesized using the Prime-Script RT Master Mix (Promega M-MLV, USA) according to the manufacturer’s instructions. qRT-PCR was performed using SYBR Green Realtime PCR Master Mix (TAKARA, Japan) in an LightCycler 480 II machine (Roche). The primer sequences for CENPK used in this study were as follows: sense: 5′- ATGGTACTGTCCACTAAGGAGTC-3′, antisense: 5′- TGTTCATCCAACCACCGTTGT-3′. The primer sequences for GAPDH as control used in this study were as follows: sense: 5′-TGACTTCAACAGCGACACCCA-3′, antisense: 5′- CACCCTGTTGCTGTAGCCAAA-3′.
Protein extraction and western blot analysis
Total proteins were extract from each sample with RIPA buffer (Beyotime, China) containing protease and phosphatase inhibitors (1mM, Cell Signaling Technology, USA) on ice for 30 min. Cell lysates were centrifuged for 15 minutes at 14,000 × g and 4 °C. The supernatant that containing proteins were retained. Then the concentrations of proteins were determined using a BCA Protein Assay Kit (Thermo Scientific, USA). Protein samples (30 μg) were loaded on 10% SDS-PAGE gel (Life Technology, USA) and transferred to a 0.45-μm PVDF membrane (Millipore, USA). The membrane was then incubated with the primary antibody at 4 °C for 24 h. CENPK was detected with a mouse polyclonal anti-CENPK antibody (BIOSS, China). GAPDH or β-actin (BIOSS, China) was used as an internal control. The secondary antibody used was goat anti-mouse IgG-HRP (BIOSS, China). The band was imaged and analyzed using a visual imaging system (SageCaptureTM MiniChemi, Sage, China).
Cell proliferation assay with high content living cell imaging system and CCK-8 analysis
Cells that infected with lentivirus had green fluorescence. The number of cells in 96-well plates of different groups were counted, analyzed and taken photos with High content living cell imaging system (Thermo, USA) for consecutive 3–5 days. The cell proliferation assays were performed with CCK-8 assay. After the cells were infected with shRNA lentivirus for 5 days, 2000 cells were seeded into the 96-well plate. From the next day, 10 microliters CCK-8 reagent (Dojindo, Kumamoto, Japan) solution were added to each well and incubate for 2 hours. Then the absorbance at 450 nm was measured with Microplate Reader (Tecan infinite). Each well has 3–5 duplicates.
Cell colony-formation assay
Anchorage independent growth was detected using a colony formation assay. Briefly, after the cells were transfected with shRNA lentivirus, cells of each experimental group in logarithmic growth stage were digested with trypsin and 1000 cells were loaded per well in six-well plates. After cells were cultured for approximately 14 days or the cell number in single colony were over 50, cells were fixed with 4% paraformaldehyde, and stained with 0.1% crystal violet (Sigma, St. Louis, MO). Then the cell colonies were photographed with inverted microscope (Zeiss Vert A1). Finally, the total number of colonies was counted and analyzed. Each well was performed in triplicate.
Cell wound-healing and cell migration assays
For cell wound-healing assay, after the cells infected with shRNA lentivirus containing GFP for 48 hours, cells were digested and counted, 50000 cells per well were seeded in the 96-well plate. The next day, make a wound using tips in each well. Wash the plate with PBS for 2–3 times, and change the media with new media with 1% FBS. Each well was performed in triplicate. Then the wells were observed under the high content living cell imaging system (Thermo, USA) for 48 hours. The cell migration rate was calculated with the following formula: [ (cell area at 48 hour) − (cell area at 0 hour) ]/[1 − (cell area at 0 hour) ]. For cell migration assays, 105cells per well were seeded in the upper chamber of 24-well Boyden chambers (Corning, Corning, NY, with 8-μm pores and without matrigel) with FBS free media, meantime, fill in 30%FBS media in the lower chamber. Sixteen hours later, the upper chamber were taken out, non-invasive cells were scraped off with cotton swabs, the invasive cells were fixed in the 4% Paraformaldehyde and stained with 0.5% crystal violet solution for 10 min at room temperature. Finally, the cells were observed under microscope (Zeiss Vert A1).
The experiments were performed in triplicate.
Cell cycle and apoptosis assay with flow cytometry
For cell cycle assay, after cells were infected with shRNA lentivirus for 5 days, cells in different groups were trypsined and seeded in the 6-cm dish and cultured until 80% confluent, trypsined and collected the cells, stained with PI (Sigma, USA) and detected with flow cytometry (Millipore, Guava easyCyte HT), data were analyzed with ModFit software. For cell apoptosis assay, according to the instruction of apoptosis kit (eBioscience, China), the cells were stained with Annexin V-APC and detected with flow cytometry (BD, USA). There were three repetitions in each group.
Animal study and in vivo tumorigenic assays
BALB/c-nu/nu female mice (6 weeks old) were purchased from the Shanghai SLAC Laboratory Animal company (Shanghai, China). All animals were housed and maintained under SPF (specific pathogen-free) environment. All animal experiments were approved by the Experimental animal ethics committee in Gansu Provincial Hospital. After gastric cancer cells MKN45 were infected with shCENPK-1 successfully, 4 × 106 cells in 200 μL PBS were injected subcutaneously at dorsal region of nude mice. Mice with xenograft tumor were observed with mall animal in vivo imaging system (Perkin Elmer Lumina LT, USA). Moreover, tumor length and width were measured with Vernier caliper every three days, and tumor volumes (cm3) were calculated with the formula: 3.14/6 × Length × Width2. Finally, tumor weight (mg) was recorded after mice were euthanasia.
Co-IP and LC-MS
The potential interacting proteins of target gene CENPK were screened by Co-IP (Co-Immunoprecipitation) and LC-MS (Liquid Chromatograph-Mass Spectrometry) assay. First, By PCR technology, the gene sequence of 3 × FLAG tag is fused to the 5′- end of the target gene CENPK to generate fusion gene of 3 × Flag with CENPK, then the fusion gene was inserted into the corresponding Lentivirus Expression Vector to prepare the expression plasmid: P3 × FLAG-target; after that, Lentivirus empty vector and P3 × Flag with CENPK plasmid separately were transfected with auxiliary plasmid into 293T cells to harvest lentivirus particles, namely Lenti-control and Lenti-3 × FLAG-CENPK. At last, the gastric cancer cells AGS were infected with the above lentivirus, and puromycin were used to screen stable cell lines, which were named as control cell (NC) and 3 × Flag-CENPK cell (OE). Second, after gel running, the cell proteins were collected and Co-IP assay were done with anti-flag antibody, the gel band were cut and the peptide segment were extracted for LC-MS test and analysis to screen the interaction proteins. Third, we further verified the Co-IP binding through the specific antibody of the candidate interaction proteins. Finally, for functional recovery experiment, we constructed the interference vectors targeting the screened positive interaction proteins XRCC5 and JUP, the gastric cancer cell AGS were then infected with both shCENPK-1 lentivirus and XRCC5 or JUP overexpression lentivirus. Furthermore, the cell proliferation abilities were studied with High content living cell imaging system (Thermo, USA) for 5 consecutive days. Meantime, the cell migration abilities were also observed to verify the functional recovery effect of XRCC5 on the target gene CENPK. At last, recovery of cellular function by XRCC5 in gastric cancer cell lines supported by immunoblotting to show expression of XRCC5, CENPK and other genes related to apoptosis (p53, Bcl-2, Cleaved Caspase-3) to clarify the detailed mechanism.
Statistic analysis
There were at least three individuals in each group and each experiments were conducted in triplicate. All the data are presented as mean values ± standard deviation (SD). The results were statistic with ANOVA test, t test or Pearson Chi-square. P < 0.05 was considered statistically significant. All data were analyzed using SPSS version 21.0 for Windows (IBM, Armonk, NY, USA).
Results
The CENPK expression level is higher in several gastric cancer cell lines
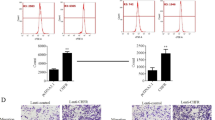
To display the differential expression of CENPK in TCGA (The Cancer Genome Atlas) database more intuitively, 26 original data of TCGA RNA SEQ samples were presented by a line chart (Fig. 1a) and the form of histogram (Fig.1b). Comparing with the adjacent normal tissue, CENPK expression in Stomach adenocarcinoma (STAD) was significantly upregulated. The mean FC (ratio of cancer samples to adjacent samples) of CENPK was 3.699, and the P value is 2.31E-04. We also detected CENPK mRNA expression level in GC (gastric cancer) cell lines by qRT-PCR (Fig. 1c). The value was represented with ∆Ct (CENPK-ACTB). When ∆CT≤12, the mRNA expression was high; When 12≤ ∆CT≤ 16, the gene mRNA expression in the cell was medium; When ΔCT value≥16, the mRNA gene expression was low. Among five GC cell lines (AGS, BGC-823, MGC80-3, SGC-7901 and MKN45), CENPK mRNA expression was significantly increased.
The CENPK expression level in Gastric cancer cell lines and tissues. a The fold line diagram of CENPK mRNA expression amount in TCGA RNA SEQ samples between cancer and normal tissues. The vertical axis is each sample, and the horizontal axis is the cancer tissue and adjacent normal tissue of the sample. Each sample has a fold line, and the trend of the broken line shows the change trend of the gene in all samples. b The FC (ratio of cancer samples to adjacent samples) histogram of CENPK mRNA expression amount in TCGA RNA SEQ samples. The vertical axis is logFC (take the logarithm value with 2 as the bottom for FC), and the horizontal axis is different samples. c CENPK mRNA expression by qRT-PCR in five gastric cancer cells. The value was represented with ∆Ct (CENPK-ACTB). ACTB was utilized as internal reference. All the reactions were run in triplicate. Data are represented as mean±SD. d Representative IHC staining for CENPK and Ki-67 in human gastric cancer and normal tissues (scale bar: 100 μm for 20× and 50 μm for 40× for objective lense). e qRT-PCR assay for knockdown efficiency of shCENPK-1 and shCENPK-2 in AGS cells. f qRT-PCR assay for knockdown efficiency of shCENPK-1 and shCENPK-2 in MKN 45 cells. g Western blot assay for knockdown efficiency of shCENPK-1 and shCENPK-2 in AGS cells. h Western blot assay for knockdown efficiency of shCENPK-1 and shCENPK-2 in MKN45 cells. All the reactions were run in triplicate. *P < 0.05 and **P < 0.01 comparing with shCtrl group
Prognostic and clinic pathological significance of CENPK in gastric cancer
To explore whether CENPK expression has significant difference between each group associated with clinic pathological features of Gastric cancer cases, IHC staining was performed in 63 human Gastric cancer samples, and high CENPK protein expression was detected in 33.3% (21/63) of those GC samples (Fig. 1d). Also, we did the immunohistochemistry of human tissues with anti-ki-67. As shown in Fig. 1d, with the increasing IHC score, the gastric cancer tissue exhibited more ki-67 expression (brown particles in the nuclear) and increased CENPK staining, which signified that CENPK might promote proliferation in gastric cancer.
Meanwhile, to compare if the CENPK expression level (percentage in each group) has significant difference between each group under the listed clinical pathological parameters in Table 1, we use SPSS to manipulate the Pearson Chi-square test or Fisher's exact test. P value was calculated for statistical difference analysis to evaluate if there was significant difference between each groups. As shown in Table 1, under histologic differentiation, the percentage of high CENPK expression in “well or moderate” group was 35% (14/40), in “poor” group was 65.2% (15/23); while the percentage of low CENPK expression in in “well or moderate” group was 15% (26/40), in “poor” group was 34.8%(8/23). After Pearson Chi-square test, the P value for these groups percentage was 0.021, which means CENPK expression level had difference in these two groups under histologic differentiation. The results were similar in two groups (I-II and III-IV) under TNM stage, with the P value 0.014. However, in other groups, P > 0.05, which meaned that the CENPK expression levels had no differences in groups under Sex, Age, Serum CEA Level, Lymph vascular invasion, as well as perineal invasion. Taken together, CENPK expression level has significant difference in different groups under GC histologic differentiation and TNM stage. Poor differentiation and III-IV stage had more percentages of high CENPK expression.
Knockdown of CENPK suppressed cancer cell proliferation, motility and migration in vitro
To investigate the potential role of gene CENPK on gastric cancer cells, we further explored cell proliferation and migration abilities when knockdown gene CENPK in cancer cells. First, gastric cancer cells (AGS and MKN45) were infected with lentivirus shCENPK-1 and shCENPK-2 to knock down CENPK. The knockdown effect in cells of each shRNA was confirmed by qRT-PCR (Fig. 1e, f) and western blot (Fig. 1g, h). The results of qRT-PCR showed that after lentivirus infection, comparing with control group, the mRNA expression of CENPK gene in AGS and MKN45 cells decreased (P < 0.05), and the knockdown efficiency of shCENPK-1 in AGS cells reached 55.4%; ShCENPK-2 knockdown efficiency reached 58.0%. In MKN45 cells, compared with control group, the knockdown efficiency of CNEPK gene in shCENPK-1 and shCNEPK-2 group were 49.1% and 67.4% separately (Fig. 1e, f).
Second, cells infected with shCENPK-1 and shCENPK-2 were observed under High Content Living Cell Imaging System (HCS) for 5 consecutive days (Fig. 2a, b). According to the statistics, comparing to the shCtrl group, cell count assays of AGS (Fig. 2c) and MKN45 cells (Fig. 2d), as well as cell count/fold assay (ratio of cells number in CENPK knockdown group to that in control group) of AGS (Supplementary Figure 1a) and MKN45cells (Supplementary Figure 1b) showed that CENPK knock down markedly inhibited cell growth (P <0.01). For example, cell count/fold on the fifth day of AGS cells in shCtrl group was 8.179, while the cell count/fold in shCNEPK-1 and shCENPK-2 groups were only 3.625 and 1.505 separately. Cell count/fold on the fifth day of MKN45 cells in shctrl group was 4.02, while that in shCNEPK-1 and shCNEPK-2 in groups were only 1.92 and 1.10 separately (supplementary Figure 1a, b).
Proliferation assay, CCK-8 and colony formation assay in cells with CENPK knockdown. Panel a and b Gastric cancer cells AGS and MKN45 infected with shCENPK-1 and shCENPK-2 were observed by HCS for 5 consecutive days. The original size of HSC image is about 100 times as the same resolution (96dpi, 1 µm × 1 µm) of the microscope. c and d Cell count assays of AGS and MKN45cells of HSC showed that CENPK knock down markedly inhibited cell growth vs. shCtrl group. e, f CCK-8 proliferation assays in AGS cells and MKN45 cells. g and h Colony formation assays of AGS cells. All the reactions were run in triplicate. Data are represented as mean±SD, and analyzed with t Test in SPSS software. *P < 0.05 and **P < 0.01 comparing with shCtrl group
Third, we determined cancer cell proliferation by CCK-8 and colony formation assays. Absorbance was read at 450nm using Tecan Microplate Reader. The value of OD450 can reflect the number of viable cells. The CCK-8 assays showed that CENPK knockdown could inhibit proliferation in both AGS (Fig. 2e) and MKN45 cells (Fig. 2f) comparing with the control group. The ratio of OD450 between the experimental group and the control group (OD450/fold) was also calculated. The value of OD450/fold on the fifth day in the control group in AGS cells increased by 7.0874 times, while the value of OD450/fold in shCENPK-1 and shCENPK-2 groups increased by only 4.5378 times and 2.4322 times, which were significantly decreased (Supplementary Figure 1c). The same results could be seen in MKN45 cells (Supplementary Figure 1d). Similar results were obtained in colony formation assays (Fig. 2g, h). Comparing with control group, when knocking down CENPK, the number of clone decreased significantly.
Moreover, scratch assay demonstrated that cell lines (AGS and MKN45) display less motility when knockdown CENPK comparing with control group (Fig. 3a and b). The average 24-hour migration rate of the control group (shCtrl) in AGS cells was 90.18%, while the average migration rate of shCENPK-1 and shCENPK-2 groups were only 65.08% and 52.12% separately (Fig. 3c). The experimental results of MKN45 cells were the same as those of AGS cells. The average 48-hour migration rate of the control group (shCtrl) in MKN45 cells was 33.93%, while the migration rate of the experimental group (shCENPK-1 and shCENPK-2) were only 8.60% and 3.18% separately (Fig. 3d).
Scratch assay and migration assay of gastric cancer cells with CENPK knockdown. a and b Scratch assay pictures of AGS and MKN45 cells. The same field were photographed at certain time points under HSC. The original size of HSC image was about 100 times as the same resolution (96 dpi, 1μm×1μm) of the microscope. c and d The quantification for migration rate in scratch assay in AGS and MKN45 cells. Bars represent the migration rate of each group, expressed as the value calculated with the formula noted in the materials and methods. e The Transwell migration assays in AGS cells with knocking down CENPK. The magnification is 100×. f The quantification of migratory cells per field in AGS cells. Cells from three random were counted with Image J software. Error bars represented the SE for n = 5 experiments carried out in triplicate. The P values were obtained using the Student’s paired t-Test, two-tailed. *P < 0.05 and **P < 0.01 comparing with shCtrl group. g Western blot analysis of EMT marker (E-cadherin, Snail, MMP9 and Slug) for MKN45 cells. β-actin as internal control
Furthermore, Transwell migration assays also showed that knocking down CENPK significantly attenuated the migratory abilities of AGS cells (Fig. 3e). The average number of cells migrated in the control group (shCtrl) was 169, while the average number of cells migrated in the experimental group (shCENPK-1 and shCENPK-2) were only 18 and 9, which were significantly decreased (P < 0.01) (Fig. 3f). At the same time, we calculated the migration fold change. The average migration fold of shCENPK-1 group was 0.11 (18/169) and shCENPK-2 group was 0.05 (9/169) (Supplementary Figure 1e).
Meanwhile, we did western blot analysis of several EMT (Epithelial Mesenchymal Transition) markers (E-cadherin, Snail, MMP9 and Slug) for MKN45 cells. The expression of protein Snail, MMP9 and Slug were decreased in CENPK knockdown groups (Fig. 3g), which proved that CENPK gene knockdown inhibited the process of EMT. Taken together, these findings suggested that knockdown of CENPK suppressed cancer cell growth, proliferation, motility and migration abilities in gastric cancer cells.
CENPK knockdown resulted in G1 arrest in gastric cancer cells
To investigate the reason of cell proliferation inhibition after knocking down CENPK, we did cell cycle analysis with flow cytometry. Remarkably, in MKN45 cells, we found that cells percentage of the G1 phase in both shCENPK-1 and shCENPK-2 groups were increased comparing with that of shCtrl group (P < 0.05), the percentage of cells in the S phase in shCENPK-2 transfected group were significantly decreased (P < 0.01), meanwhile, the percentage of cells in the G2/M phase in both shCENPK-1 and shCENPK-2 transfected groups decreased too (P < 0.05) (Fig. 4a, b). Together, the data showed that CENPK knockdown inhibited gastric cancer cell growth by arresting cells in G1 phase.
Cell cycle and apoptosis analysis with flow cytometry. a The cell cycle analysis in MKN 45 cells. b The percentage (%) of cells in G1, S and G2/M phases in shCENPK-1, shCENPK-2 groups and control groups. Panel c and d The apoptosis analysis on gastric cancer cells AGS and MKN45 cells. e, f The quantification of apoptosis rate for AGS and MKN45 cells. All experiments carried out in triplicate. *P < 0.05 and **P < 0.01 comparing with shCtrl group with t-Test in SPSS software
CENPK knockdown increased apoptosis rate in gastric cancer cells
To further study how CENPK knocking down inhibited cell proliferation and growth, we did cell apoptosis analysis in gastric cancer cells. We noted that the apoptosis rate of AGS cells in shCENPK-1 and shCENPK-2 groups were 10.50% and 12.27% respectively, which were greatly increased than shCtrl group 2.62% (P < 0.01) (Fig. 4c and e); For MKN45 cells, the apoptosis rate in shCENPK-1 and shCENPK-2 groups were 7.03% and 48% respectively, which were greatly increased than shCtrl group 1.23% (P < 0.01) (Fig. 4d and f). These findings suggested that CENPK knockdown could increase apoptosis rate in gastric cancer cells, which meaned that gene CENPK might decrease gastric cancer cells apoptosis, then leading to the cancer cell over-proliferation.
Knockdown of CENPK suppressed tumorigenesis in vivo
To further delineate whether knockdown of CENPK could impact gastric cancer growth in vivo, MKN45 cells (4 × 106) infected with shCENPK-1 and shCtrl were injected subcutaneously into 6-week female BALB/c-nu/nu mice. First, we assessed tumor volume with Small Animal in Vivo Imaging System. We found that the xenograft tumor in CENPK knockdown group had less total and average radiant efficiency than NC group (Fig. 5a–d). After the animals were euthanized, mice injected with cells that knocking down CENPK (KD) had smaller tumor volume and tumor weight comparing with control group (NC). The average tumor volume of the animals in the KD group on the 15th day was 485.92 cm3, while in the control group on the 15th day tumor had grown to 772.48 cm3. The average tumor weight in KD group after euthanasia was 0.508g, while tumor weight in NC group was 0.806g (Fig. 5e–h). We also did experiment of mice tumor histology. We did both HE staining and anti-ki-67 staining for the tumor tissue. As shown in Fig. 5i, ki-67 expression (brown particles in the nuclear) in KD group was less than that in NC group. The results indicated that CENPK knockdown could reduce the proliferation of gastric cancer in vivo. In conclusion, we showed that knockdown of CENPK could suppressed tumorigenesis and proliferation in vivo.
Knockdown of CENPK suppressed tumorigenesis in vivo. Panel a and b Mice tumor volume of CENPK knockdown group (KD) and control group (NC)were assessed with Small Animal in Vivo Imaging System. c The average radiant efficiency ([p/s/cm2/s]/[μW/cm2]) calculated by the Animal in Vivo Imaging System. d The total radiant efficiency ([p/s/cm2/s]/[μW/cm2]) calculated by the animal in vivo imaging system. For c and d, animal number 1–10 represented the control group, while number 11–20 represented KD group. e, f The mice and tumor display after animals were euthanized. g Tumor volume (cm3) was measured at certain time point, and calculated with the formula: 3.14/6 × Length × Width2. h Tumor average weight after the xenograft tumor were Peeled off. i HE staining and anti-ki-67 staining for the mice tumor tissue (scale bar: 100 μm for 20× and 50 μm for 40× for objective lense). Each group had ten mice, and The P values were obtained using the Student’s paired t-Test, two-tailed. *P < 0.05 and **P < 0.01 comparing with NC group
Co-IP combined with LC-MS analysis to screen the potential proteins interacting with CENPK
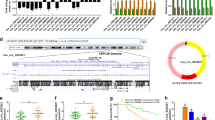
To explore the downstream mechanism of CENPK in gastric cancer, we did Co-IP (Co-Immunoprecipitation) and LC-MS (Liquid Chromatograph-Mass Spectrometry) analysis to screen the potential proteins that interacting with gene CENPK. First, we use FLAG antibody to detect the protein of 3 × FLAG-CENPK protein both in NC group and OE group, we noted that in OE group, there was the protein of FLAG tag, which proved the successful gene fusion of FLAG and CENPK (Fig. 6a). Second, we did Co-IP pre experiment, the result showed that after Co-IP with FLAG-beads, flag tag was detected in OE group, indicating that flag tag can be normally exposed, recognized by anti-flag antibody, and target protein can be enriched through IP (Fig. 6b). Third, we cultured and collected more cells to do the formal Co-IP experiments, SDS-PAGE electrophoresis and Coomassie brilliant blue staining results of Co-IP protein samples demonstrated the protein of 3 × FLAG-CENPK after Co-IP (Fig. 6c). Forth, the gel strips were cut off, the protein in each sample strip was digested into peptide segments with trypsin, then each peptide sample was identified by LC-MS. The original mass spectrometry files of each sample were analyzed with PD/MASCOT software to obtain the protein identification. The total spectra, PSM (Peptide Spectrum Match), Peptides, and Protein groups were shown in Supplementary figure 2a, comparing with the NC group, there were 328 different interacting proteins with CENPK (Supplementary table 2). Fifth, the list of proteins specifically identified by OE group was analyzed with IPA bioinformatics. The interaction between gene and target gene CENPK was predicted. Finally, we did Co-IP experiments again with several protein antibodies predicted in the IPA analysis (JUP, ACTA2, XRCC5, PARP1, HNRNPD, PARP1 and HSPB1) to verify the potential proteins that might interact with target protein CENPK. Remarkably, we found that in the IP group, comparing with NC group, XRCC5 and JUP had significant more expression (Fig. 6d). In IP:Flag group, when overexpression CENPK(OE), the immunoblotting results indicated that XRCC5 and JUP in OE group were stronger than that in NC group. We conclude that CENPK might interact with XRCC5 and JUP.
Screening of CENPK interaction complexes by Co-IP combined with LC-MS, and Functional Recovery Study of interaction proteins of CENPK. a The Flag expression in AGS cells infected with Lenti-control (NC) and Lenti-3 × FLAG-CENPK (OE)was detected by Western blot, β-actin as internal control. b Co-IP pre-experiment. ‘IP: flag’ represented cells proteins were detected with anti-flag with western blot after immunoprecipitated with FLAG, ‘Input’ represent whole cell lysate before immunoprecipitation. c SDS-PAGE electrophoresis and Coomassie brilliant blue staining results of Co-IP protein samples in the formal Co-IP experiments. The red arrow pointed to the 3×FLAG-CENPK protein. Marker from top to bottom: 250 kDa, 150 kDa, 100 kDa, 75 kDa, 50 kDa, 37 kDa, 25 kDa, 20 kDa and 15 kDa. d Verification of several proteins that might interact with target protein CENPK with western blot assay. In the IP group, comparing with NC group, XRCC5 and JUP had significant more expression, which might be interacted with CENPK. e The AGS cell proliferation abilities recovery assay was studied with HCS for 5 consecutive days. Cells infected with shCENPK-1 lentivirus to knockdown CENPK (KD) were green, while infected with shJUP (OE1) and shXRCC5 (OE2) lentivirus to make them overexpression were red. The original size of HSC image was about 100 times as the same resolution(96dpi, 1 μm×1 μm)of the microscope. f The quantification of AGS cell number under HSC in proliferation abilities recovery assay. g MTT assay of AGS cells to study the recovery function of over expression of XRCC5 to CENPK knockdown (KD). OE represented overexpression of XRCC5. h Transwell migration assay to further study the recovery function of over expression of XRCC5 to CENPK knockdown. i The quantification of migratory cells per field in each group. KD represented CENPK knockdown, OE represented overexpression of XRCC5. j The immunoblotting to show expression of XRCC5, CENPK and other genes related to apoptosis (P53, Bcl-2, Cleaved Caspase-3). OE represented overexpression of XRCC5. All experiments were carried out in triplicate. *P < 0.05 and **P < 0.01 comparing with control group
Functional recovery study of interaction proteins to CENPK
To further test whether XRCC5 and JUP indeed interacted with CENPK, we did Functional recovery study. First, gastric cancer cells AGS were infected with shCENPK-1 lentivirus (GFP) to knockdown CENPK (KD), then infected with overexpression shJUP or shXRCC5 lentivirus (red) to make them overexpression (OE1 and OE2), the cell proliferation abilities were studied with HCS for 5 consecutive days. Compared with NC+NC group, the proliferation of KD+NC group decreased significantly, which was consistent with the previous results (P < 0.05); However, compared with KD+NC group, after over expression JUP or XRCC5, in KD+OE1 and KD+OE2 groups, the cell number significantly increased, so the proliferation ability of AGC cells was recovered (P < 0.05) (Fig. 6e and f). We also calculated the ratio of cell number to the previous day (cell number/fold). At the fifth day, comparing with the fold value in KD+NC group (2.13), the value of KD+OE1 and KD+OE2 groups were 6.33 and 8.79, respectively (Supplementary figure 2b).
Second, we did MTT assay to further study the recovery function of over expression of XRCC5 to AGS cells. Compared with NC+NC group (the OD490 value at the fifth day was 0.589), the cell activity of KD+NC group decreased (the OD490 value at the fifth day was 0.113, P < 0.05), which was consistent with the previous results. However, compared with KD+NC group, the cell activity of KD+OE group increased (the OD490 value at the fifth day was 0.314, P < 0.05), indicating that XRCC5 gene overexpression restored the proliferation function of CENPK knockdown (Fig. 6g). We also calculated the OD490/fold, which is the ratio of OD490 value to the previous day, the results were consistent (Supplementary figure 2c).
Moreover, Similar results were obtained in Transwell migration assay (Fig. 6h and i). The average migratory cells per field were 236 in control group, when knocking down CENPK (KD+NC), the average migratory cells were decreased to 17 (P < 0.05), then after XRCC5 over expression ((KD+OE), the average migratory cells were increased to 125 (P < 0.05). Similar results could be seen when migration fold change were calculated (Supplementary figure 2d). The result indicated that XRCC5 gene overexpression (KD+OE) restored the migration ability of CENPK knockdown.
Finally, to clarify the recovery of cellular function by XRCC5 in gastric cancer cell lines and the detailed mechanism, we did immunoblotting to show expression of XRCC5, CENPK and other genes related to apoptosis (P53, Bcl-2, Cleaved Caspase-3) when knocking down CENPK (KD) and overexpression XRCC5(OE). As shown in Fig. 6j, in NC+KD group, CENPK and XRCC5 decreased, however, the P53, Bcl-2 and Cleaved Caspase-3 increased, which indicated that knocking down CENPK could contribute to apoptosis, the results were similar with former experiments in vitro (Fig.4c and e). However, when overexpression XRCC5, besides the CENPK and XRCC5 increased, the expression of P53, Bcl-2 and Cleaved Caspase-3 were significantly decreased, which demonstrated CENPK and XRCC5 might interact to promote gastric cancer through decreasing apoptosis.
In conclusion, we showed that XRCC5 gene overexpression can restore the proliferation and migration abilities of gastric cancer cells that knocking down CENPK, thereby proved that XRCC5 might interacted with CENPK together to play the function of promoting gastric cancer growth and metastasis.
Discussion
In the present study, we showed that CENPK expression is upregulated in several gastric cancer cell lines and tissue samples. Clinically, under statistical analysis, CENPK expression level has significant difference in different groups under GC histologic differentiation (P = 0.021) and TNM stage (P = 0.014). Poor differentiation and III-IV stage had more percentages of high CENPK expression, higher expression of CENPK and ki-67 was observed in poor differentiation cancer tissues, which give us a clue that CENPK might be a novel oncogene for gastric cancer. In the following research, we found that knocking down CENPK significantly inhibited gastric cancer cell proliferation and migration, while promoted the apoptosis rate and cause the cell cycle arrested in the G1 stage. Moreover, knocking down CENPK also inhibited tumor growth in vivo. These results indicated that CENPK plays a critical role in gastric cancer initiation and progression and deserve further study.
Centromeric proteins can be divided into five groups of sub complex, CENP-H/I/K/M is one of them. According to the bioinformatics analysis, CENP-K and CENP-H are both enriched in coiled-coils regions, and they may form coiled-coils in the kinetochores, therefore might directly involve in the attachment between kinetochores and spindle microtubules [12]. Similar research also reported that CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates [13], and structural analysis of fungal CENP-H/I/K homologs reveals a conserved assembly mechanism underlying proper chromosome alignment [14]. Through bioinformatics analysis combine with experiments, some research had demonstrated that CENPK was associated with cancer cell proliferation, such as thyroid carcinoma [15], lung adenocarcinoma [16]. Some reports proved that CENPK was also a prognostic and predictive biomarkers in small cell lung cancer [17] and conjunctival melanoma [5] , as well as a novel therapeutic targets for triple negative breast cancer [18]. Our research investigated the role of CENPK in gastric cancer and proved first time the CENPK also upregulated in gastric cancer, knocking down CENPK could inhibit cell growth both in vitro and in vivo, could induced the apoptosis and cause the cell arrest at G1 stage.
The most encouraging finding in this study is that we showed for the first time that XRCC5 might be a novel interaction protein with CENPK. There were few researches about the downstream genes or proteins of CENPK, most of them demonstrated that knockdown of CENPK inhibited cell proliferation and facilitates apoptosis though regulating YAP1 [10], or by activating AKT/P53 [19], or JAK/STAT3 signaling pathway [20], these were the well-known signal pathway. However, through Co-IP and LC-MS, our study showed for the first time that XRCC5 (X-Ray Repair Cross Complementing 5) might be a novel interaction protein with CENPK. The Protein encoded by XRCC5 gene is the 80-kilodalton subunit of the Ku heterodimer protein which is also known as ATP-dependent DNA helicase II or DNA repair protein XRCC5. Ku is the DNA-binding component of the DNA-dependent protein kinase, and it functions together with the DNA ligase IV-XRCC4 complex in the repair of DNA double-strand break by non-homologous end joining and the completion of V(D)J recombination events [21]. This gene functionally complements Chinese hamster xrs-6, a mutant defective in DNA double-strand break repair and in ability to undergo V(D)J recombination [22]. XRCC5 polymorphisms might be related with breast cancer risk [23] and chemotherapy response [24]. Also, higher XRCC5 expression can predict poor prognosis in glioblastoma [25].
According to our research results and relevant reports, we speculated that in gastric cancer cells, the expression of CENPK gene expression increased, and the kinetochore assembly caused by centromeric protein complex is accelerated, resulting in accelerated cell cycle (especially the G1 phase), and excessive cell proliferation. Therefore, the expression of XRCC5 gene related to DNA damage repair is also increased, resulting in abnormal DNA damage repair, which may lead to reduced apoptosis and promote EMT, thus lead to the development of gastric cancer. However, more specific mechanism between CNEPK and XRCC5 needs to be deeply explored in the future.
There are some limitations for this paper. First, JUP function with CENPK deserve further investigate. In this study, by Co-IP and LC-MS, we identified 328 proteins that may interact with CENPK, and 35 potential proteins that may interact with CENPK to promote the cancer cell proliferation and migration were selected by bioinformatics. Due to conditions, we only selected ACTA2, HNRNPD, HSPb1, JUP, PARP1 and XRCC5 proteins for subsequent Co-IP verification. The validation results showed that both JUP (junction plakoglobin) and XRCC5 interacted with the target protein CENPK (Fig. 6d). In functional recovery study by HCS, the reduced proliferation ability caused by CENPK knockdown could be recovered both by over-expression of JUP and XRCC5 gene (Fig. 6e and f). Therefore, JUP may also be a potential CENPK interacting gene and a potential target for the treatment of gastric cancer. It is worthy of further exploration and research. In addition, how CENPK and XRCC5 interact to promote the proliferation and migration of gastric cancer cells need to be further investigated. It is unclear how XRCC5 plays a role in the function of CENPK in decreasing apoptosis and promoting G1 phase. Finally, if CENPK also play important role in the DNA double-strand break repair process and how XRCC5 function in gastric cancer have not identified, we will further study the underlying mechanism in the future work.
Conclusions
Collectively, our results revealed that CENPK was upregulated in two kinds of gastric cancer cells and tissues. CENPK expression level has significant difference in groups under GC histologic differentiation and TNM stage. Poor differentiation and III-IV stage had more percentages of high CENPK expression. Knockdown CENPK in two kinds RNA sequences silencing (shCENPK-1 and shCENPK-2) could both significantly inhibit cell proliferation and migration via interacting with XRCC5. Our findings not only demonstrated the oncogenic role of CENPK in gastric cancer, but also highlighted the potential downstream interaction protein of CENPK.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A, Global cancer statistics,. GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. https://doi.org/10.3322/caac.21492.
Digklia A, Wagner AD. Advanced gastric cancer: Current treatment landscape and future perspectives. World J Gastroenterol. 2016;22(8):2403–14. https://doi.org/10.3748/wjg.v22.i8.2403.PubMedPMID:26937129;PubMedCentralPMCID:PMC4768187.
Hoischen C, Yavas S, Wohland T, Diekmann S. CENP-C/H/I/K/M/T/W/N/L and hMis12 but not CENP-S/X participate in complex formation in the nucleoplasm of living human interphase cells outside centromeres. PloS one. 2018;13(3): e0192572. https://doi.org/10.1371/journal.pone.0192572.PubMedPMID:29509805;PubMedCentralPMCID:PMC5839545.
Zhang Z, Bellini D, Barford D. Crystal structure of the Cenp-HIKHead-TW sub-module of the inner kinetochore CCAN complex. Nucleic Acids Res. 2020;48(19):11172–84. https://doi.org/10.1093/nar/gkaa772.PubMedPMID:32976599;PubMedCentralPMCID:PMC7641736.
Wolf J, Auw-Haedrich C, Schlecht A, Boneva S, Mittelviefhaus H, Lapp T, et al. Transcriptional characterization of conjunctival melanoma identifies the cellular tumor microenvironment and prognostic gene signatures. Sci Rep. 2020;10(1):17022. https://doi.org/10.1038/s41598-020-72864-0.PubMedPMID:33046735;PubMedCentralPMCID:PMC7550331.
Liu Y, Xiong S, Liu S, Chen J, Yang H, Liu G, et al. Analysis of Gene Expression in Bladder Cancer: Possible Involvement of Mitosis and Complement and Coagulation Cascades Signaling Pathway. J Comput Biol. 2020;27(6):987–98. https://doi.org/10.1089/cmb.2019.0237.
Gong W, Guo P, Liu L, Guan Q, Yuan Z. Integrative Analysis of Transcriptome-Wide Association Study and mRNA Expression Profiles Identifies Candidate Genes Associated With Idiopathic Pulmonary Fibrosis. Front Genet. 2020;11: 604324. https://doi.org/10.3389/fgene.2020.604324.PubMedPMID:33362862;PubMedCentralPMCID:PMC7758323.
Lee Y, Huang C, Lin D, Chang W, Lee K. Overexpression of centromere protein K (CENPK) in ovarian cancer is correlated with poor patient survival and associated with predictive and prognostic relevance. PeerJ. 2015;3: e1386.
Wang H, Liu W, Liu L, Wu C, Wu W, Zheng J, et al. Overexpression of centromere protein K (CENP-K) gene in hepatocellular carcinoma promote cell proliferation by activating AKT/TP53 signal pathway. Oncotarget. 2017;8(43):73994–4005.
Wang J, Li H, Xia C, Yang X, Dai B, Tao K, et al. Downregulation of CENPK suppresses hepatocellular carcinoma malignant progression through regulating. Onco Targets. 2019;12:869–82. https://doi.org/10.2147/OTT.S190061.
Ma J, Chen X, Lin M, Wang Z, Wu Y, Li J. Bioinformatics analysis combined with experiments predicts CENPK as a potential prognostic factor for lung adenocarcinoma. Cancer Cell Int. 2021;21(1):65. https://doi.org/10.1186/s12935-021-01760-y.PubMedPMID:33478508;PubMedCentralPMCID:PMC7818917.
Qiu S, Wang J, Yu C, He D. CENP-K and CENP-H may form coiled-coils in the kinetochores. Sci China Ser C, Life Sci. 2009;52(4):352–9. https://doi.org/10.1007/s11427-009-0050-3 (PubMed PMID: 19381461).
Cheeseman IM, Hori T, Fukagawa T, Desai A. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol Biology Cell. 2008;19(2):587–94. https://doi.org/10.1091/mbc.e07-10-1051.PubMedPMID:18045986;PubMedCentralPMCID:PMC2230600.
Hu L, Huang H, Hei M, Yang Y, Li S, Liu Y, et al. Structural analysis of fungal CENP-H/I/K homologs reveals a conserved assembly mechanism underlying proper chromosome alignment. Nucleic Acids Res. 2019;47(1):468–79. https://doi.org/10.1093/nar/gky1108.PubMedPMID:30407575;PubMedCentralPMCID:PMC6326798.
Li Q, Liang J, Zhang S, An N, Xu L, Ye C. Overexpression of centromere protein K (CENPK) gene in differentiated thyroid carcinoma promote cell proliferation and migration. Bioengineered. 2021;12(1):1299–310. https://doi.org/10.1080/21655979.2021.1911533 (PubMed PMID: 33904381).
Wang Y, Wang Y, Ren C, Wang H, Zhang Y, Xiu Y. Upregulation of centromere protein K is crucial for lung adenocarcinoma cell viability and invasion. Adv Clin Exp. 2021;30(7):691–9. https://doi.org/10.17219/acem/133820 (PubMed PMID: 34118147).
Gao C, Gu X, Chen Y, Zhou M, Jiang F, Zheng S. Identification of potential prognostic and predictive biomarkers for immune-checkpoint inhibitor response in small cell lung cancer. Med Sci Monit: Int Med J Exp Clin Res. 2021;27: e932275. https://doi.org/10.12659/MSM.932275.PubMedPMID:34719665;PubMedCentralPMCID:PMC8570048.
Komatsu M, Yoshimaru T, Matsuo T, Kiyotani K, Miyoshi Y, Tanahashi T, et al. Molecular features of triple negative breast cancer cells by genome-wide gene expression profiling analysis. Int J Oncol. 2013;42(2):478–506. https://doi.org/10.3892/ijo.2012.1744 (PubMed PMID: 23254957).
Wang H, Liu W, Liu L, Wu C, Wu W, Zheng J, et al. Overexpression of centromere protein K (CENP-K) gene in hepatocellular carcinoma promote cell proliferation by activating AKT/TP53 signal pathway. Oncotarget. 2017;8(43):73994–4005. https://doi.org/10.18632/oncotarget.18172.PubMedPMID:29088763;PubMedCentralPMCID:PMC5650318.
Jia B, Dao J, Han J, Huang Z, Sun X, Zheng X, et al. LINC00958 promotes the proliferation of TSCC via miR-211-5p/CENPK axis and activating the JAK/STAT3 signaling pathway. Cancer Cell Int. 2021;21(1):147. https://doi.org/10.1186/s12935-021-01808-z.PubMedPMID:33658048;PubMedCentralPMCID:PMC7931557.
Singh JK, Smith R, Rother MB, de Groot AJL, Wiegant WW, Vreeken K, et al. Zinc finger protein ZNF384 is an adaptor of Ku to DNA during classical non-homologous end-joining. Nature Commun. 2021;12(1):6560. https://doi.org/10.1038/s41467-021-26691-0.PubMedPMID:34772923;PubMedCentralPMCID:PMC8589989.
Monferran S, Muller C, Mourey L, Frit P, Salles B. The Membrane-associated form of the DNA repair protein Ku is involved in cell adhesion to fibronectin. J Mol Biol. 2004;337(3):503–11. https://doi.org/10.1016/j.jmb.2004.01.057 (PubMed PMID: 15019772).
Yu LX, Liu LY, Xiang YJ, Wang F, Zhou F, Huang SY, et al. XRCC5/6 polymorphisms and their interactions with smoking, alcohol consumption, and sleep satisfaction in breast cancer risk: a Chinese multi-center study. Cancer Med. 2021;10(8):2752–62. https://doi.org/10.1002/cam4.3847.PubMedPMID:33734613;PubMedCentralPMCID:PMC8026916.
Zuo K, Yuan X, Liang X, Sun X, Liu S, Connell PP, et al. qRT-PCR-based DNA homologous recombination-associated 4-gene score predicts pathologic complete response to platinum-based neoadjuvant chemotherapy in triple-negative breast cancer. Breast Cancer Res Treat. 2021. https://doi.org/10.1007/s10549-021-06442-x (PubMed PMID: 34797456).
Lee IN, Yang JT, Huang C, Huang HC, Wu YP, Chen JC. Elevated XRCC5 expression level can promote temozolomide resistance and predict poor prognosis in glioblastoma. Oncology Lett. 2021;21(6):443. https://doi.org/10.3892/ol.2021.12704.PubMedPMID:33868481;PubMedCentralPMCID:PMC8045174.
Acknowledgments
This work was supported by National Natural Science Foundation of China (81902364) , NHC Key Laboratory of Diagnosis and Therapy of Gastrointestinal Tumor Gansu Provincial Hospital (NLDTG2020022), Natural Science Foundation of Gansu Province (20JR10RA403), Open Fund of Gansu Key Laboratory of Surgical Tumor Molecular Diagnosis and Precision Therapy (2019GSZDSYS06), Special project of the central government to guide local scientific and Technological Development (2021-07).
Funding
Gansu Science and Technology Department, 20JR10RA403, Hongwei Tian, Young Scientists Fund, 81902364, Fang Wang, NHC Key Laboratory of Diagnosis and Therapy of Gastrointestinal Tumor Gansu Provincial Hospital (NLDTG2020022), Hongwei Tian, Open Fund of Gansu Key Laboratory of Surgical Tumor Molecular Diagnosis and Precision Therapy (2019GSZDSYS06), Hongwei Tian, Special project of the central government to guide local scientific and Technological Development (2021-07) Fang Wang.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests. Fang Wang, and Hongwei Tian contributed equally to this work.
Ethical approval
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and later versions.
Informed consent
Informed consent to be included in the study, or the equivalent, was obtained from all patients.
Human and animal rights
All institutional and national guidelines for the care and use of laboratory animals were followed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tian, H., Wang, F., Deng, Y. et al. Centromeric protein K (CENPK) promotes gastric cancer proliferation and migration via interacting with XRCC5. Gastric Cancer 25, 879–895 (2022). https://doi.org/10.1007/s10120-022-01311-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-022-01311-y