Abstract
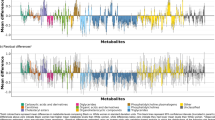
Complex multi-omics effects drive the clustering of cardiometabolic risk factors, underscoring the imperative to comprehend how individual and combined omics shape phenotypic variation. Our study partitions phenotypic variance in metabolic syndrome (MetS), blood glucose (GLU), triglycerides (TG), high-density lipoprotein cholesterol (HDL-C), and blood pressure through genome, transcriptome, metabolome, and exposome (i.e., lifestyle exposome) analyses. Our analysis included a cohort of 62,822 unrelated individuals with white British ancestry, sourced from the UK biobank. We employed linear mixed models to partition phenotypic variance using the restricted maximum likelihood (REML) method, implemented in MTG2 (v2.22). We initiated the analysis by individually modeling omics, followed by subsequent integration of pairwise omics in a joint model that also accounted for the covariance and interaction between omics layers. Finally, we estimated the correlations of various omics effects between the phenotypes using bivariate REML. Significant proportions of the MetS variance were attributed to distinct data sources: genome (9.47%), transcriptome (4.24%), metabolome (14.34%), and exposome (3.77%). The phenotypic variances explained by the genome, transcriptome, metabolome, and exposome ranged from 3.28% for GLU to 25.35% for HDL-C, 0% for GLU to 19.34% for HDL-C, 4.29% for systolic blood pressure (SBP) to 35.75% for TG, and 0.89% for GLU to 10.17% for HDL-C, respectively. Significant correlations were found between genomic and transcriptomic effects for TG and HDL-C. Furthermore, significant interaction effects between omics data were detected for both MetS and its components. Interestingly, significant correlation of omics effect between the phenotypes was found. This study underscores omics’ roles, interaction effects, and random-effects covariance in unveiling phenotypic variation in multi-omics domains.





Similar content being viewed by others
Data availability and software
The genotype and phenotype data of the UK Biobank used in this study are publicly accessible through procedures described on its webpage (http://www.ukbiobank.ac.uk/using-the-resource). We confirm that the data supporting the findings of this study are available within the article and its supplementary files. The source code for MTG version 2.22 is publicly available in https://sites.google.com/view/s-hong-lee-homepage/mtg2.
References
Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA et al (2009) Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation 120(16):1640–1645
Ambroselli D, Masciulli F, Romano E, Catanzaro G, Besharat ZM, Massari MC et al (2023) New advances in metabolic syndrome, from prevention to treatment: the role of diet and food. Nutrients 15(3):640
Auwerx C, Sadler MC, Woh T, Reymond A, Kutalik Z, Porcu E (2023) Exploiting the mediating role of the metabolome to unravel transcript-to-phenotype associations. Elife 12:e81097
Baik I, Shin C (2008) Prospective study of alcohol consumption and metabolic syndrome. Am J Clin Nutr 87(5):9
Barbeira AN, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, Torres JM et al (2018) Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun 9(1):1825
Barbeira AN, Pividori M, Zheng J, Wheeler HE, Nicolae DL, Im HK (2019) Integrating predicted transcriptome from multiple tissues improves association detection. PLoS Genet 15(1):e1007889
Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R et al (2017) Heart Disease and Stroke Statistics-2017 update: A report from the American Heart Association. Circulation 135(10):e146–e603
Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K et al (2018) The UK Biobank resource with deep phenotyping and genomic data. Nature 562(7726):203–209
Carioca AAF, Steluti J, Carvalho AM, Silva AM, Silva I, Fisberg RM et al (2021) Plasma metabolomics are associated with metabolic syndrome: a targeted approach. Nutrition 83:111082
Che T, Yan C, Tian D, Zhang X, Liu X, Wu Z (2021) The association between sleep and metabolic syndrome: a systematic review and meta-analysis. Front Endocrinol (lausanne) 12:773646
Chen M, Yang Z, Gan H, Wang Y, Li C, Gao Y (2022) Investigation into potential mechanisms of metabolic syndrome by integrative analysis of metabolomics and proteomics. PLoS One 17(7):e0270593
Cho JH, Ko J, Lim ST (2021) Relationship between metabolic syndrome and moderate-to-vigorous physical activity among adults 18 years old and over. PLoS One 16(10):e0258097
Despres JP, Lemieux I, Bergeron J, Pibarot P, Mathieu P, Larose E et al (2008) Abdominal obesity and the metabolic syndrome: contribution to global cardiometabolic risk. Arterioscler Thromb Vasc Biol 28(6):1039–1049
Drouard G, Ollikainen M, Mykkanen J, Raitakari O, Lehtimaki T, Kahonen M et al (2022) Multi-omics integration in a twin cohort and predictive modeling of blood pressure values. OMICS 26(3):130–141
Eicher T, Kinnebrew G, Patt A, Spencer K, Ying K, Ma Q et al (2020) Metabolomics and multi-omics integration: a survey of computational methods and resources. Metabolites 10(5):202
Fahed G, Aoun L, Bou Zerdan M, Allam S, Bou Zerdan M, Bouferraa Y et al (2022) Metabolic syndrome: updates on pathophysiology and management in 2021. Int J Mol Sci 23(2):786
Fathi DB (2018) The investigations of genetic determinants of the metabolic syndrome. Diabetes Metab Syndr 12(5):783–789
Gamazon ER, Wheeler HE, Shah K, Mozaffari SV, Aquino-Michaels K, Carroll RJ et al (2015) PrediXcan: Trait mapping using human transcriptome regulation. Nat Genet 47:1091–1098
Gurka MJ, Golden SH, Musani SK, Sims M, Vishnu A, Guo Y et al (2017) Independent associations between a metabolic syndrome severity score and future diabetes by sex and race: the Atherosclerosis Risk In Communities Study and Jackson Heart Study. Diabetologia 60(7):1261–1270
Hasin Y, Seldin M, Lusis A (2017) Multi-omics approaches to disease. Genome Biol 18(1):83
Huang S, Chaudhary K, Garmire LX (2017) More is better: recent progress in multi-omics data integration methods. Front Genet 8:84
Jolliffe IT (1982) A note on the use of principal components in regression. J R Stat Soc C 31(3):300–303
Julkunen H, Cichonska A, Slagboom PE, Wurtz P, Nightingale Health UKBI (2021) Metabolic biomarker profiling for identification of susceptibility to severe pneumonia and COVID-19 in the general population. Elife 10:e63033
Kraja AT, Vaidya D, Pankow JS, Goodarzi MO, Assimes TL, Kullo IJ et al (2011) A bivariate genome-wide approach to metabolic syndrome: STAMPEED consortium. Diabetes 60(4):1329–1339
Kristiansson K, Perola M, Tikkanen E, Kettunen J, Surakka I, Havulinna AS et al (2012) Genome-wide screen for metabolic syndrome susceptibility loci reveals strong lipid gene contribution but no evidence for common genetic basis for clustering of metabolic syndrome traits. Circ Cardiovasc Genet 5(2):242–249
Lanktree MB, Hegele RA (2017) Metabolic syndrome. Genomic and precision medicine, pp 283–299
Lee SH, van der Werf JHJ (2006) An efficient variance component approach implementing an average information REML suitable for combined LD and linkage mapping with a general complex pedigree. Genet Sel Evol 38(1):25–43
Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR (2012) Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics 28(19):2540–2542
Lind L (2019) Genome-wide association study of the metabolic syndrome in UK Biobank. Metab Syndr Relat Disord 17(10):505–511
Lind L, Sundstrom J, Elmstahl S, Dekkers KF, Smith JG, Engstrom G et al (2022) The metabolomic profile associated with clustering of cardiovascular risk factors—a multi-sample evaluation. PLoS One 17(9):e0274701
Louca P, Tran TQB, Toit CD, Christofidou P, Spector TD, Mangino M et al (2022) Machine learning integration of multimodal data identifies key features of blood pressure regulation. EBioMedicine 84:104243
Mohr S, Liew CC (2007) The peripheral-blood transcriptome: new insights into disease and risk assessment. Trends Mol Med 13(10):422–432
Ni G, van der Werf J, Zhou X, Hypponen E, Wray NR, Lee SH (2019) Genotype-covariate correlation and interaction disentangled by a whole-genome multivariate reaction norm model. Nat Commun 10(1):2239
Qiao Z, Sidorenko J, Revez JA, Xue A, Lu X, Parna K et al (2023) Estimation and implications of the genetic architecture of fasting and non-fasting blood glucose. Nat Commun 14(1):451
Rattray NJW, Deziel NC, Wallach JD, Khan SA, Vasiliou V, Ioannidis JPA et al (2018) Beyond genomics: understanding exposotypes through metabolomics. Hum Genom 12(1):4
Revelas M, Thalamuthu A, Zettergren A, Oldmeadow C, Najar J, Seidu NM et al (2023) High polygenic risk score for exceptional longevity is associated with a healthy metabolic profile. Geroscience 45:399–413
Rutten-Jacobs LC, Larsson SC, Malik R, Rannikmae K, MEGASTROKE consortium; International Stroke Genetics Consortium et al (2018) Genetic risk, incident stroke, and the benefits of adhering to a healthy lifestyle: cohort study of 306 473 UK Biobank participants. BMJ 363:k4168
Saklayen MG (2018) The global epidemic of the metabolic syndrome. Curr Hypertens Rep 20(2):12
Santiago-Rodriguez TM, Hollister EB (2021) Multi ’omic data integration: a review of concepts, considerations, and approaches. Semin Perinatol 45(6):151456
Sun YV, Hu YJ (2016) Integrative analysis of multi-omics data for discovery and functional studies of complex human diseases. Adv Genet 93:147–190
Tropf FC, Lee SH, Verweij RM, Stulp G, van der Most PJ, de Vlaming R et al (2017) Hidden heritability due to heterogeneity across seven populations. Nat Hum Behav 1(10):757–765
Vahabi N, Michailidis G (2022) Unsupervised multi-omics data integration methods: a comprehensive review. Front Genet 13:854752
van Buuren S, Groothuis-Oudshoorn K (2011) mice: Multivariate Imputation by Chained Equations in R. J Stat Softw 45(3):67
van Walree ES, Jansen IE, Bell NY, Savage JE, de Leeuw C, Nieuwdorp M et al (2022) Disentangling genetic risks for metabolic syndrome. Diabetes 71(11):2447–2457
Wu Q, Li J, Sun X, He D, Cheng Z, Li J et al (2021) Multi-stage metabolomics and genetic analyses identified metabolite biomarkers of metabolic syndrome and their genetic determinants. EBioMedicine 74:103707
Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88(1):76–82
Zhou X, Lee SH (2021) An integrative analysis of genomic and exposomic data for complex traits and phenotypic prediction. Sci Rep 11(1):21495
Zhou X, Im HK, Lee SH (2020) CORE GREML for estimating covariance between random effects in linear mixed models for complex trait analyses. Nat Commun 11(1):4208
Acknowledgements
L.D.A acknowledges the Australian Government Research Training Program (RTP) and University of South Australia for funding his PhD scholarship. We obtained data from UK Biobank under the reference number 14575. We would like to thank all participants and staff of the UK Biobank for their valuable contributions. We would like to thank the Statistical Genetics Research Group for their advice during the conception and planning stages of the study. The analyses were performed using computational resources provided by the Australian Government through Gadi under the National Computational Merit Allocation Scheme(NCMAS), and HPCs (Statgen servers) managed by UniSA IT. The data used to impute gene expression levels were retrieved from GTEx V8 through the GTEx portal (www.gtexportal.org) and dbGaP accession number is phs000424.v8.p2.
Funding
This research is supported by the Australian Research Council (DP190100766).
Author information
Authors and Affiliations
Contributions
SHL and LDA conceived the idea, directed the study, performed the data management, and wrote the manuscript; LDA conducted the analyses; SHL supervised the study. SHL and LDA wrote the first draft of the manuscript. All the authors provided critical feedback and suggestions. All the authors critically reviewed the manuscript and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Amente, L.D., Mills, N.T., Le, T.D. et al. Unraveling phenotypic variance in metabolic syndrome through multi-omics. Hum. Genet. 143, 35–47 (2024). https://doi.org/10.1007/s00439-023-02619-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-023-02619-0