Abstract
Aims/hypothesis
Correctly diagnosing MODY is important, as individuals with this diagnosis can discontinue insulin injections; however, many people are misdiagnosed. We aimed to develop a robust approach for determining the pathogenicity of variants of uncertain significance in hepatocyte nuclear factor-1 alpha (HNF1A)-MODY and to obtain an accurate estimate of the prevalence of HNF1A-MODY in paediatric cases of diabetes.
Methods
We extended our previous screening of the Norwegian Childhood Diabetes Registry by 830 additional samples and comprehensively genotyped HNF1A variants in autoantibody-negative participants using next-generation sequencing. Carriers of pathogenic variants were treated by local healthcare providers, and participants with novel likely pathogenic variants and variants of uncertain significance were enrolled in an investigator-initiated, non-randomised, open-label pilot study (ClinicalTrials.gov registration no. NCT04239586). To identify variants associated with HNF1A-MODY, we functionally characterised their pathogenicity and assessed the carriers’ phenotype and treatment response to sulfonylurea.
Results
In total, 615 autoantibody-negative participants among 4712 cases of paediatric diabetes underwent genetic sequencing, revealing 19 with HNF1A variants. We identified nine carriers with novel variants classified as variants of uncertain significance or likely to be pathogenic, while the remaining ten participants carried five pathogenic variants previously reported. Of the nine carriers with novel variants, six responded favourably to sulfonylurea. Functional investigations revealed their variants to be dysfunctional and demonstrated a correlation with the resulting phenotype, providing evidence for reclassifying these variants as pathogenic.
Conclusions/interpretation
Based on this robust classification, we estimate that the prevalence of HNF1A-MODY is 0.3% in paediatric diabetes. Clinical phenotyping is challenging and functional investigations provide a strong complementary line of evidence. We demonstrate here that combining clinical phenotyping with functional protein studies provides a powerful tool to obtain a precise diagnosis of HNF1A-MODY.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.

Introduction
Monogenic diabetes affects up to 6.5% of autoantibody-negative Norwegian children with diabetes aged under 15 years [1, 2], with most cases explained by individuals carrying a single, heterozygous gene variant impairing insulin secretion [3]. The biggest subtype, MODY [4, 5], can be caused by a dysfunction in one of 11 genes, of which the gene encoding hepatocyte nuclear factor-1 alpha (HNF1A) serves as the largest contributor in Norway [1]. Hepatocyte nuclear factor-1 alpha (HNF1A)-MODY follows an autosomal dominant inheritance pattern with affected individuals usually found in at least three generations. Individuals typically present with non-ketotic hyperglycaemia and are diagnosed with diabetes in childhood or adolescence [6,7,8,9].
MODY is frequently misdiagnosed as cases can be difficult to recognise. This can be explained by overlapping phenotypes with other types of diabetes [1, 10, 11], incomplete penetrance and clinical features that are not always consistent with the classical MODY criteria [12,13,14,15,16,17]. The autosomal dominant inheritance pattern can also be masked by insufficient clinical information. For this reason, it is essential that systematic screening is performed to uncover misclassified cases that are not picked up on clinical suspicion.
Monogenic diabetes provides a good example where precision diagnostics, including in-depth pathophysiological knowledge, can pave the way for targeted therapy. In diagnosed individuals, sulfonylurea, acting on KATP channels in pancreatic beta cells, and thereby increasing insulin secretion specifically, improves metabolic control and quality of life [18,19,20,21,22]. Identifying children misclassified as having type 1 diabetes is therefore of great importance, as they can discontinue painful and cumbersome insulin injections, and early detection, when endogenous insulin secretion is highest, translates to a better prognosis when switching to sulfonylurea [23].
The availability of effective treatments has expedited the use of high-throughput sequencing for comprehensive genotyping. While a powerful approach, variant interpretation can still pose a significant challenge. Gene variants are sorted into five categories; pathogenic, likely pathogenic (LP), variant of uncertain significance (VUS), likely benign (LB) or benign [24,25,26,27]. Many variants fall into the VUS category due to either limited or conflicting evidence pointing in both benign and pathogenic directions. Furthermore, many disease-causing HNF1A variants remain as VUSs due to lack of clinical data and limited or ambiguous results from molecular studies [4]. In the case of a VUS, the individual is left with an unclear molecular diagnosis, and clinicians may misinterpret this result as non-pathogenic. In truth, a classification of VUS simply indicates that more evidence is needed to determine whether or not the variant is pathogenic.
We previously performed a systematic screening of the Norwegian Childhood Diabetes Registry (NCDR), which covers >99% of paediatric diabetes cases in Norway [28], to identify carriers of HNF1A variants in autoantibody-negative children [1, 10]. That study gave a lower estimate of prevalence of HNF1A-MODY of 2.2% (excluding VUSs) and an upper estimate of 4.1% (including VUSs). By determining which VUSs are in fact pathogenic, we can not only provide a better treatment for these individuals, but also obtain a more accurate estimate of the true prevalence.
In this study, we extend the scope of our previous search to identify individuals with novel LP variants and VUSs. By combining functional studies of the HNF1A protein with advanced physiological and clinical treatment response data, including a sulfonylurea switch trial, we demonstrate that we can robustly classify these variants. This comprehensive approach can be used to systematically screen and identify HNF1A-MODY and certain other MODY types, and lead to increased diagnostic precision in children with diabetes.
Methods
We designed an investigator-initiated, non-randomised, open-label pilot study to explore sulfonylurea (ClinicalTrials.gov registration no. NCT04239586) as treatment for HNF1A-MODY in individuals with pathogenic variants, LP variants or VUSs in HNF1A. All participants or their legal guardians gave written informed consent, and our research was carried out in accordance with the Declaration of Helsinki. The Western Norway Regional Ethics Committee approved the study (no. 2009/2080 and no. 2018/2388).
Individuals from genetic screening of the NCDR
NCDR is a Norwegian population-based registry of children aged 0–18 years with newly diagnosed diabetes. By a recent analysis pairing data with the Patient Registry of Norway (http://www.kvalitetsregistre.no, in Norwegian), it was found to cover >99% of paediatric cases and 97.3% of those aged 0–18 years with diabetes in Norway. It is representative for the Norwegian childhood diabetes population regarding sex, ethnicity, age, and regional and socioeconomic factors. At diagnosis, information on ethnicity is collected in addition to clinical and biochemical data, as well as blood samples. From our updated screen we included 830 additional children from the NCDR (diagnosed between March 2015 and July 2017). In addition to the 3882 individuals included in the study by Johansson et al [1], this gave us a total of 4712 participants. Of these, 623 participants had negative autoantibodies (cut-offs: GADA antibody index <0.08 or 1.0 U/ml, IA-2A antibody index <0.1 or 1.0 U/ml) and were subjected to next-generation sequencing (NGS) on an Illumina MiSeq (Illumina, San Diego, CA, USA) sequencer at Hudson Alpha Institute for Biotechnology (Huntsville, AL, USA). The panel set-up included a total of 13 MODY genes (covering all MODY genes recognised to date except RFX6): HNF1A, GCK, HNF4A, HNF1B, INS, ABCC8, KCNJ11, BLK, CEL, NEUROD1, KLF11, PAX4 and PDX1. Detailed information is provided in Johansson et al [1]. As eight participants were excluded from further analysis due to poor DNA quality, the remaining 615 participants (462 [2015] + 153 [2017]) were screened for HNF1A variants interpreted according to the ClinGen Monogenic Diabetes Expert Panel specifications to the American College of Medical Genetics and Genomics/the Association for Molecular Pathology (ACMG/AMP) guidelines [27] (electronic supplementary material [ESM] Table 1). Variants classified as benign or LB by the latest guidelines were excluded. For carriers of well-characterised pathogenic variants, we advised local healthcare providers on recommended treatment (sulfonylurea) with a later follow-up by interview in 2021.
Carriers of novel LP variants and VUSs were invited to participate in a sulfonylurea switch trial and physiological assessments in the form of an OGTT. Participants with very low stimulated C-peptide levels on a test meal (C-peptide <0.1 nmol/l) were excluded. One positive control participant, a newly diagnosed carrier of the pathogenic HNF1A variant p.Arg203His treated with insulin, was included for comparison in the switch trial.
Treatment trial of participants carrying novel LP variants and VUSes
Re-examination of autoantibody status
At the time of admittance, autoantibodies were measured once again, this time also adding ZnT8A to the two previously tested (cut-offs: GADA: <5 U/ml, IA-2A: <7.5 U/ml, ZnT8A: <15 U/ml).
OGTT
Short-acting insulin was discontinued 2 h prior to testing, while long-acting insulin was stopped from 22:00 hours the previous day. Following an overnight fast, 75 g (1.75 g/kg if <40 kg) of glucose (equal to 82.5 g monohydrated) mixed with 200–300 ml of water was ingested at time 0 and plasma was collected at −15, 0, 30, 60, 90 and 120 min for assays of glucose, C-peptide and insulin.
Sulfonylurea treatment trial
Glipizide (2.5 mg), a second-generation sulfonylurea, was administered at the 2 h mark of the OGTT. The dose was adjusted gradually according to response over the next few days. Response to sulfonylurea was carefully monitored during the admission. Response was defined as maintaining euglycaemia (finger stick capillary glucose 4.0 to <10.0 mmol/l or 70 to <180 mg/dl) without using insulin within the maximal recommended dosage according to the Norwegian Medicines Agency (https://legemiddelverket.no, accessed multiple times, first time 18 April 2017). Measures were taken to avoid hypoglycaemic incidents during the OGTT and after the switch to sulfonylurea (information, glucose sensors, carbohydrate-rich food at hand, frequent testing of capillary and plasma glucose, and admittance to hospital). In the case of symptomatic hypoglycaemia or random measurement <3.0 mmol/l, testing was to be stopped, and glucose given orally or intravenously. To avoid any risk of ketoacidosis, ketones (capillary and urine) were measured at the 120 min mark of the OGTT, and rechecked until negative in participants with positive measurements. In these participants, reinstatement of insulin was considered early. The study physician (PS) carried out weekly telephone consultations in the first weeks after discharge. Responders were invited for another assessment, with a second OGTT, at a minimum of 3 months after discharge. Participants were instructed to take glipizide before this second assessment.
Functional studies
Construction of HNF1A variant plasmids for expression analysis
HNF1A variants were constructed using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA) and variant-specific primers. Individual HNF1A variants were introduced into the wild-type HNF1A cDNA isoform A (NCBI NM_000545.6, with substitutions; c.51C>G, p.Leu17= and c.79A>C, p.Ile27Leu) in the pcDNA 3.1/HisC vector. In the transactivation assays, the firefly luciferase reporter vector pGL3-RA (Promega, Madison, WI, USA), containing the rat albumin promoter cloned into a pGL3 vector, as well as the Renilla luciferase control vector pRLSV40 (Promega) were used as described previously [24, 26].
Luciferase reporter assays and protein abundance
HeLa cells were cultured and transiently transfected with wild-type or variant HNF1A cDNA, together with the firefly reporter plasmid and the Renilla reporter (pRLSV40) as an internal control. Luciferase activity was measured 24 h post transfection using the Dual-Luciferase Assay System (Promega) on a Centro XS3 LB 960 luminometer (Berthold Technologies, Germany). Next, the level of HNF1A protein expression in the wild-type and variants was assessed in cell lysates obtained for the transactivation assays. In short, 20 µl of cell lysates was subjected to SDS-PAGE and immunoblotting using antibodies against HNF1A (Cell Signaling, Beverly, MA, USA) and α-tubulin (Abcam, Cambridge, MA, USA), with α-tubulin as a loading control. For the five pathogenic variants, these variants have been comprehensively investigated previously [25, 26, 29, 30] and thus only the luciferase assay was reinvestigated.
Nuclear fractionation
Nuclear and cytosolic fractions were isolated from transiently transfected HeLa cells as previously performed [31]. Total protein in each fraction was measured using Bradford reagent (Thermo Fisher, Waltham, MA, USA) and 8 µg of total protein from each fraction was subjected to SDS-PAGE and immunoblotting with antibodies for HNF1A (Cell Signaling). The relative subcellular localisation was calculated by using the ratios of HNF1A with the respective nuclear (topoisomerase IIα [Abcam]) and cytosolic (α-tubulin [Abcam]) markers.
DNA binding studies
The electrophoretic mobility shift assays were carried out as previously described [31]. Briefly, equal protein amounts of nuclear fractions from transiently transfected HeLa cells were incubated together with a cyanine 5-labelled oligonucleotide (Sigma Aldrich, St Louis, MO, USA), using the Odyssey EMSA buffer kit (LI-COR Biosciences, Lincoln, NE, USA) and the promoter of the rat Alb gene (5′-TGTGGTTAATGATCTACAGTTA-3′) for the binding reaction.
Statistical analysis
We present results as means ± SD and relative to wild-type HNF1A (set to 100%), unless stated otherwise. Statistical differences were analysed using two-tailed Student’s t tests with a significance level of p<0.05, using GraphPad Prism software version 8.1.1 (GraphPad Software, San Diego, CA, USA).
Results
Clinical assessments
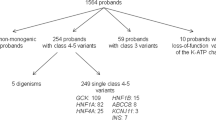
Of the 4712 children with diabetes from the NCDR, 615 were GADA and IA-2A negative and subjected to genetic screening. Of these, 19 participants were identified as carrying HNF1A variants classified as pathogenic, LP or VUS (Fig. 1). Nine carriers of eight different novel LP variants or VUSs in HNF1A were identified (Table 1) and included in the switch trial. Their diagnoses had not been confirmed but two participants were treated with sulfonylurea on clinical suspicion of MODY. The remaining ten participants carried variants already established as pathogenic and were, as expected, sensitive to diet or insulin secretagogues (ESM Table 2). Only one carrier in this group was not previously diagnosed with HNF1A-MODY.
Timeline for screening HNF1A variants in the NCDR. The NCDR was screened in 2015 and 2017, including a total of 4712 children with diabetes. Of these, 623 participants were autoantibody negative and eligible for genetic analysis, but only 615 of these samples displayed high-quality DNA or coverage. Ten participants with five different pathogenic variants were identified, and these were switched to MODY treatment (sulfonylurea) by local paediatricians. An additional nine carriers of novel LP variants or VUSs were invited to an inpatient switch trial in which insulin was replaced with sulfonylurea, with investigation of the phenotype and response to treatment. Molecular and functional investigations of participants’ HNF1A variants complemented the clinical assessments
In the treatment switch trial, six of the nine participants responded to sulfonylurea and three responded poorly to the switch, judged by a failed stimulated C-peptide test pre-admission or an insufficient response to sulfonylurea. Of the six participants with preserved insulin secretion (Fig. 2a–c), careful examination revealed co-segregation (variant and phenotype appear in the same individuals in a family) of the variant and diabetes in only three families, the rest being inconclusive due to missing clinical/family information (p.Gln561*), multiple diabetes phenotypes in the family (p.Ser451Gly) and reduced penetrance in the parent carrier (p.Thr547Argfs*5) (ESM Fig. 1). Reassessing the participants who successfully switched revealed sustained or improved HbA1c in these individuals at follow-up (ESM Fig. 2), and a participant with a continuous glucose monitor also showed improvement in time in range [32]) (ESM Fig. 3). One individual who successfully switched was lost to follow-up.
C-peptide measurements from the OGTT. (a) C-peptide levels in the variant carriers during a 2 h OGTT. Participants included in the switch trial showed heterogeneous insulin secretion. Carriers are coloured according to the response to sulfonylurea: switched (light red) or not switched (blue) vs the pathogenic control participant (dark red). The carrier of p.Pro580Leu was excluded from the switch trial and the results depicted are a stimulated C-peptide test. (b) AUC for participants during the OGTT, sorted by decreasing numbers within the groups. (c) Maximum incremental C-peptide for participants. The p.Lys222del variant carriers are siblings, and the individual with a lower insulin response (up-pointing triangles) is the female sibling detailed in Table 1
Of the three non-responsive VUS carriers, the p.Gln175Glu carrier was of South Asian heritage and overweight with an elevated insulin requirement before switch and raised HOMA-IR, indicating insulin resistance and type 2 diabetes (Table 1). In the switch trial, sulfonylurea failed as monotherapy at the maximum recommended dose and insulin was started at day 2. Oral sulfonylurea was withdrawn after only 2 weeks. The two other participants failing the switch attempt (carriers of p.Gly306Val and p.Pro580Leu) revealed almost no insulin secretion, both showing a type 1 diabetes phenotype, and one developed elevated GADA during the study period. Lineage studies revealed a lack of co-segregation, contradicting HNF1A-MODY, in these participants (ESM Fig. 1).
Functional studies
The positions of the 12 HNF1A variants (four pathogenic, two novel LP variants and six VUSs) in the HNF1A protein domain, and their effect on normal HNF1A transcriptional activity, protein abundance, nuclear localisation and DNA binding ability, are shown in Fig. 3a–e (corresponding western blots in ESM Fig. 4). The pathogenic variant p.Gly292Argfs*25 was not included in the functional investigation as it was previously shown to induce a frame shift, with a premature stop codon triggering nonsense-mediated decay of the RNA and preventing the truncated protein from being made [33]. Of the 12 remaining variants, nine (p.Pro112Leu, p.Arg131Trp, p.His143Pro, p.Arg203His, p.Lys222del, p.Arg229Gln, p.Ser451Gly, p.Thr547Argfs*5 and p.Gln561*) demonstrated reduced levels of transcriptional activity (12–57% of wild-type), while three variants (p.Gln175Glu, p.Gly306Val and p.Pro580Leu) demonstrated only mildly reduced activity (70–85% of wild-type). We observed significantly lower protein expression levels for the p.His143Pro and p.Lys222del variants (41% and 30%, respectively) compared with wild-type expression levels. These two variants also exhibited a significant reduction in nuclear protein level and severely reduced DNA binding (<10%). Co-expression of increasing quantities of p.Thr547Argfs*5 or p.Gln561* with fixed levels of wild-type HNF1A excluded a dominant negative effect for these variants (ESM Fig. 5).
Functional investigation of HNF1A VUSs, LP and pathogenic variants identified through screening of antibody-negative individuals in the NCDR. (a) Schematic illustration of the HNF1A protein with the positions of the identified variants. The dimerisation, DNA binding and transactivation domains of the HNF1A protein are highlighted. The pathogenic variant p.Gly292Argfs*25 is not included for reasons described in the main text. (b) Assessment of transcriptional activity of HNF1A protein variants using a luciferase reporter assay. HeLa cells were transiently transfected with wild-type or variant HNF1A plasmids together with the reporter plasmids encoding firefly (pGL3-RA) and Renilla (pRLSV40) luciferase. (c) Relative protein expression. HeLa cell lysates collected for the transactivation assay were analysed by SDS-PAGE and immunoblotting using HNF1A-specific antibodies. Protein levels, normalised to α-tubulin (loading control), are presented relative to wild-type levels (set as 100%). Representative western blots are shown in ESM Fig. 4a. (d) Nuclear localisation of HNF1A variants. Nuclear fractions of transiently transfected HeLa cells (wild-type or variant HNF1A plasmids) were assessed by SDS-PAGE immunoblotting. p.Leu197_Leu205del was used as a negative control [44], and topoisomerase IIα and α-tubulin were used as nuclear and cytosolic markers, respectively. The HNF1A/topoisomerase IIα and HNF1A/α-tubulin ratios were used to calculate the relative subcellular localisation of HNF1A in each compartment. Representative western blots are shown in ESM Fig. 4b. (e) DNA binding of HNF1A variants in an electrophoretic mobility shift assay. Equal amounts of HNF1A variants in nuclear fractions were incubated with a cyanine 5-labelled oligonucleotide corresponding to the HNF1A binding site and bound complexes were quantified by densiometric analysis (representative gel images are shown in ESM Fig. 4c). Measurements are given relative to wild-type levels (set as 100%) unless otherwise specified. Each bar represents the mean of nine readings ±SD; three parallel readings were conducted on three experimental days. *p<0.05, **p<0.01, ***p<0.001. Dark red bars: four pathogenic variants in participants identified in the screening study. Light red bars: novel LP variants or VUSs in successfully switched participants. Blue bars: VUSs in non-switched participants. EV, empty vector; SU, sulfonylurea; WT, wild-type
Discussion
In the present study, we show that 16 of 623 (2.6%) Norwegian children with antibody-negative diabetes (representing 0.3% of the NCDR) carry variants leading to impaired protein function of HNF1A, with phenotypic and functional investigations supporting HNF1A-MODY. In six cases, genetic screening revealed a novel LP variant or a VUS where further characterisation showed strong indications of HNF1A-MODY, with sustained metabolic control after switching to sulfonylurea. Our studies demonstrate that having an unclear genetic report entails a risk of being misdiagnosed and ineffectively treated with insulin. Screening for HNF1A-MODY, in addition to variant characterisation and a sulfonylurea trial, can correct a misdiagnosis.
Clinical phenotyping is challenging and an atypical presentation at the time of referral has contributed to monogenic diabetes being unrecognised, with people being clinically diagnosed as having type 1 diabetes and treated with insulin. In many cases, there was no family history (e.g. missing data, age-dependent penetrance, de novo mutation) to indicate autosomal dominant inheritance. Together, this highlights the complexity of clinical diagnostics and demonstrates the need for investigation by systematic screening and characterisation of carriers of novel variants or VUSs.
In agreement with previous reports on functional analyses of other variants [7, 8, 24, 25, 30], loss of transactivation in p.Thr547Argfs*5 and p.Gln561* variants cannot be explained by impaired DNA binding ability, nuclear targeting or reduced protein expression as these subanalyses are not affected. In contrast, impaired protein function in p.His143Pro and p.Lys222del might be explained by the combined effect of several mechanisms. In this study, the p.Ser451Gly variant demonstrated only moderately reduced transcriptional activity (~60%) and did not impede any other HNF1A function. Although the clinical presentation correlated with a MODY phenotype in this individual, it is unclear whether the variant alone is causing a highly penetrant diabetes, or whether the effect could be driven in a polygenic manner, as the segregation studies were inconclusive, with a high proportion of type 2 diabetes cases in non-carriers in the family.
Consistent with sulfonylurea insensitivity in p.Gln175Glu, p.Gly360Val and p.Pro580Leu, the transactivation assays demonstrated only minor reduction (70–85% of wild-type) and no indication of pathogenic variants, indicating the presence of other diabetes subtypes in the respective carriers. This illustrates an important message that pathogenicity cannot be determined for VUS carriers, and careful characterisation of the variant is needed. Removing insulin from people with atypical type 1 diabetes (negative autoantibodies) comes with a great risk of ketoacidosis, so careful intervention in a hospital setting is critical when assessing variant carriers and treatment response in paediatric patients. In these non-responding individuals, the transactivation assay of these specific variants alone pointed to other causes of diabetes.
A strength of our study is the completeness of the NCDR, making our population-based findings representative of a northern European population, like a similar recent registry study from the Nordic region [34]. Unlike studies estimating prevalence by genetically screening only referred individuals or small subsets of individuals, our study is less affected by selection bias [11, 35,36,37,38,39]. The use of a larger NGS gene panel, containing 16 genes, in addition to selected promoter regions and the inclusion of VUSs makes it unlikely that any individual was missed in our screen. Another advantage is the robust variant interpretation guidelines used in this study [27].
Because of the rarity of the disorder, the number of individuals carrying HNF1A variants is small, even in a nationwide cohort. At the time of inclusion, the NCDR only contained children under the age of 15 years, and our lower prevalence compared with similar screening studies (0.5–1.0%) is likely a reflection of our younger cohort [40, 41]. This could be addressed in future screening trials, as the age limit of the registry has now been increased to 18 years. Ideally, all variant carriers would follow the same protocol, but as the switch study was initiated years after the screening study, many carriers had already been receiving sulfonylurea. We did, however, manage to prospectively enrol one insulin-treated carrier with a pathogenic variant who was successfully switched to sulfonylurea. In our study, we simply illustrate the large variation in insulin secretion (Fig. 2). We realise that the heterogeneity and progressiveness of HNF1A-MODY makes any comparison complex. Phenotypes of diabetes tend to overlap, and many factors affect insulin secretion, importantly age and diabetes duration, as previous studies have already established [42].
As we included only autoantibody-negative individuals in the screening study, although a reasonable selection, we risked missing a few individuals, as the specificity of antibody assays is never 100% and some individuals with MODY might be autoantibody-positive with or without related autoimmune diabetes. A potential compromise could be to include individuals with low titre autoantibodies, such as in the study by Harsunen et al [34]. We did, however, investigate the autoantibody-positive VUS carriers from Johansson et al [1] (ESM Table 1), finding little indication of HNF1A-MODY in the clinical evaluation of the specific variants (p.Ser22Arg and p.Thr354Met). In further support of p.Thr354Met being benign, our previous functional investigation of the p.Thr354Met variant [25] also did not support a damaging effect on protein function. The ACMG and the AMP evidence for these two variants is provided in ESM Table 1, and functional analyses are provided in a previous publication [25] and ESM Fig. 6. The p.Ser22Arg variant was detected in two autoantibody-positive individuals, but only one gave consent to participate in the study. The consenting participant was triple autoantobody-positive (GADA/IA-2A/ZnT8 11.4/241/333 U/ml [cut-offs: GADA: <5 U/ml, IA-2A: <7.5 U/ml, ZnT8A: <15 U/ml]), revealing no glucose-stimulated endogenous insulin secretion in an OGTT (2 h serum C-peptide <0.03 nmol/l, 2 h serum glucose 23 mmol/l). The father, aged 53 years, also carries the variant but does not have diabetes. There is no other history of diabetes in the family. However, as we found a discrepancy in the clinical and functional assessments of the p.Ser22Arg variant, further investigations, including a specific dimerisation assay, would be of interest. Also, follow-up of the father regarding diabetes status in the years to come might reveal pathogenicity with a lower penetrance, and if the proband has type 1 diabetes exclusively or double diabetes (HNF1A-MODY and type 1 diabetes). Because of these uncertainties, the variant remains a VUS after this study. However, it is notable that the allele frequency of this variant in the Genome Aggregation Database (gnomAD) is higher than one would expect in a MODY-causing variant (minor allele frequency 0.013%, 37 allele counts).
Identifying and correctly diagnosing HNF1A-MODY clinically is difficult due to the challenge of phenocopies, and sulfonylurea response alone is not a suitable diagnostic marker. Our thorough approach addresses this, thus limiting the risk of misdiagnosis for carriers of HNF1A variants. First, this approach improves discovery by screening for HNF1A-MODY in high-risk populations and, second, it combines clinical and functional investigations into a robust classification scheme that increases precision in diagnosing monogenic diabetes. Specifically, we have demonstrated that functional studies aid in determining the pathogenicity of novel variants and VUSs.
In summary, we present a comprehensive method to classify paediatric cases of HNF1A-MODY, which was used to determine the pathogenicity of novel variants and VUSs. Our approach is readily extendable to variants occurring in similar diabetes-implicated transcription factors. The impact of obtaining a correct diagnosis in these individuals is considerable, permitting insulin-free treatment and the possibility of testing at-risk family members. This brings about an important change in management in these children, improving quality of life and reducing the risk of diabetes-associated late complications.
Abbreviations
- ACMG:
-
American College of Medical Genetics and Genomics
- AMP:
-
Association for Molecular Pathology
- HNF1A:
-
Hepatocyte nuclear factor-1 alpha
- LB:
-
Likely benign
- LP:
-
Likely pathogenic
- NCDR:
-
Norwegian Childhood Diabetes Registry
- NGS:
-
Next-generation sequencing
- VUS:
-
Variant of uncertain significance
References
Johansson BB, Irgens HU, Molnes J et al (2017) Targeted next-generation sequencing reveals MODY in up to 6.5% of antibody-negative diabetes cases listed in the Norwegian Childhood Diabetes Registry. Diabetologia 60(4):625–635. https://doi.org/10.1007/s00125-016-4167-1
Hanberger L, Birkebaek N, Bjarnason R et al (2014) Childhood diabetes in the Nordic countries: a comparison of quality registries. J Diabetes Sci Technol 8(4):738–744. https://doi.org/10.1177/1932296814531479
Hattersley AT, Greeley SAW, Polak M et al (2018) ISPAD Clinical Practice Consensus Guidelines 2018: the diagnosis and management of monogenic diabetes in children and adolescents. Pediatric Diabetes 19(Suppl 27):47–63. https://doi.org/10.1111/pedi.12772
Zhang H, Colclough K, Gloyn AL, Pollin TI (2021) Monogenic diabetes: a gateway to precision medicine in diabetes. J Clin Invest 131(3):e142244. https://doi.org/10.1172/jci142244
Laver TW, Wakeling MN, Knox O et al (2022) Evaluation of evidence for pathogenicity demonstrates that BLK, KLF11, and PAX4 should not be included in diagnostic testing for MODY. Diabetes 71(5):1128–1136. https://doi.org/10.2337/db21-0844
Maestro MA, Cardalda C, Boj SF, Luco RF, Servitja JM, Ferrer J (2007) Distinct roles of HNF1β, HNF1ɑ, and HNF4ɑ in regulating pancreas development, β-cell function and growth. Endocr Dev 12:33–45. https://doi.org/10.1159/000109603
Colclough K, Bellanne-Chantelot C, Saint-Martin C, Flanagan SE, Ellard S (2013) Mutations in the genes encoding the transcription factors hepatocyte nuclear factor 1 alpha and 4 alpha in maturity-onset diabetes of the young and hyperinsulinemic hypoglycemia. Hum Mutat 34(5):669–685. https://doi.org/10.1002/humu.22279
Vaxillaire M, Abderrahmani A, Boutin P et al (1999) Anatomy of a homeoprotein revealed by the analysis of human MODY3 mutations. J Biol Chem 274(50):35639–35646. https://doi.org/10.1074/jbc.274.50.35639
Fajans SS, Bell GI, Polonsky KS (2001) Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N Engl J Med 345(13):971–980. https://doi.org/10.1056/NEJMra002168
Irgens HU, Molnes J, Johansson BB et al (2013) Prevalence of monogenic diabetes in the population-based Norwegian Childhood Diabetes Registry. Diabetologia 56(7):1512–1519. https://doi.org/10.1007/s00125-013-2916-y
Shields BM, Hicks S, Shepherd MH, Colclough K, Hattersley AT, Ellard S (2010) Maturity-onset diabetes of the young (MODY): how many cases are we missing? Diabetologia 53(12):2504–2508. https://doi.org/10.1007/s00125-010-1799-4
Rubio-Cabezas O, Hattersley AT, Njolstad PR et al (2014) ISPAD Clinical Practice Consensus Guidelines 2014. The diagnosis and management of monogenic diabetes in children and adolescents. Pediatr Diabetes 15(Suppl 20):47–64. https://doi.org/10.1111/pedi.12192
Harris A, Letourneau LR, Greeley SAW (2018) Monogenic diabetes: the impact of making the right diagnosis. Curr Opin Pediatr 30(4):558–567. https://doi.org/10.1097/mop.0000000000000643
Shepherd M, Hattersley AT (2004) “I don’t feel like a diabetic any more”: the impact of stopping insulin in patients with maturity onset diabetes of the young following genetic testing. Clin Med (Lond) 4(2):144–147. https://doi.org/10.7861/clinmedicine.4-2-144
Stanik J, Dusatkova P, Cinek O et al (2014) De novo mutations of GCK, HNF1A and HNF4A may be more frequent in MODY than previously assumed. Diabetologia 57(3):480–484. https://doi.org/10.1007/s00125-013-3119-2
Romuld IB, Kalleklev TL, Molnes J, Juliusson PB, Njølstad PR, Sagen JV (2021) Impact of overweight on glucose homeostasis in MODY2 and MODY3. Diabet Med 38(10):e14649. https://doi.org/10.1111/dme.14649
Mirshahi UL, Colclough K, Wright CF et al (2022) Reduced penetrance of MODY-associated HNF1A/HNF4A variants but not GCK variants in clinically unselected cohorts. Am J Hum Genet 109:2018–2028. https://doi.org/10.1016/j.ajhg.2022.09.014
Gloyn AL, Pearson ER, Antcliff JF et al (2004) Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med 350(18):1838–1849. https://doi.org/10.1056/NEJMoa032922
Pearson ER, Flechtner I, Njolstad PR et al (2006) Switching from insulin to oral sulfonylureas in patients with diabetes due to Kir6.2 mutations. N Engl J Med 355(5):467–477. https://doi.org/10.1056/NEJMoa061759
Pearson ER, Starkey BJ, Powell RJ, Gribble FM, Clark PM, Hattersley AT (2003) Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet 362(9392):1275–1281. https://doi.org/10.1016/S0140-6736(03)14571-0
Sagen JV, Raeder H, Hathout E et al (2004) Permanent neonatal diabetes due to mutations in KCNJ11 encoding Kir6.2: patient characteristics and initial response to sulfonylurea therapy. Diabetes 53(10):2713–2718. https://doi.org/10.2337/diabetes.53.10.2713
Bowman P, Sulen A, Barbetti F et al (2018) Effectiveness and safety of long-term treatment with sulfonylureas in patients with neonatal diabetes due to KCNJ11 mutations: an international cohort study. Lancet Diabetes Endocrinol 6(8):637–646. https://doi.org/10.1016/s2213-8587(18)30106-2
Shepherd MH, Shields BM, Hudson M et al (2018) A UK nationwide prospective study of treatment change in MODY: genetic subtype and clinical characteristics predict optimal glycaemic control after discontinuing insulin and metformin. Diabetologia 61:2520–2527. https://doi.org/10.1007/s00125-018-4728-6
Malikova J, Kaci A, Dusatkova P et al (2020) Functional analyses of HNF1A-MODY variants refine the interpretation of identified sequence variants. J Clin Endocrinol Metab 105(4):dgaa051. https://doi.org/10.1210/clinem/dgaa051
Althari S, Najmi LA, Bennett AJ et al (2020) Unsupervised clustering of missense variants in HNF1A using multidimensional functional data aids clinical interpretation. Am J Hum Genet 107(4):670–682. https://doi.org/10.1016/j.ajhg.2020.08.016
Najmi LA, Aukrust I, Flannick J et al (2017) Functional investigations of HNF1A identify rare variants as risk factors for type 2 diabetes in the general population. Diabetes 66(2):335–346. https://doi.org/10.2337/db16-0460
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424. https://doi.org/10.1038/gim.2015.30
Lanzinger S, Zimmermann A, Ranjan AG et al (2022) A collaborative comparison of international pediatric diabetes registries. Pediatr Diabetes 23(6):627–640. https://doi.org/10.1111/pedi.13362
Bjørkhaug L, Ye H, Horikawa Y, Søvik O, Molven A, Njølstad PR (2000) MODY associated with two novel hepatocyte nuclear factor-1ɑ loss-of-function mutations (P112L and Q466X). Biochem Biophys Res Commun 279(3):792–798. https://doi.org/10.1006/bbrc.2000.4024
Bjorkhaug L, Sagen JV, Thorsby P, Sovik O, Molven A, Njolstad PR (2003) Hepatocyte nuclear factor-1ɑ gene mutations and diabetes in Norway. J Clin Endocrinol Metab 88(2):920–931. https://doi.org/10.1210/jc.2002-020945
Kaci A, Keindl M, Solheim MH, Njolstad PR, Bjorkhaug L, Aukrust I (2018) The E3 SUMO ligase PIASγ is a novel interaction partner regulating the activity of diabetes associated hepatocyte nuclear factor-1ɑ. Sci Rep 8(1):12780. https://doi.org/10.1038/s41598-018-29448-w
Battelino T, Danne T, Bergenstal RM et al (2019) Clinical targets for continuous glucose monitoring data interpretation: recommendations from the international consensus on time in range. Diabetes Care 42(8):1593–1603. https://doi.org/10.2337/dci19-0028
Thomas H, Badenberg B, Bulman M et al (2002) Evidence for haploinsufficiency of the human HNF1ɑ gene revealed by functional characterization of MODY3-associated mutations. Biol Chem 383(11):1691–1700. https://doi.org/10.1515/bc.2002.190
Harsunen M, Kettunen JLT, Härkönen T et al (2023) Identification of monogenic variants in more than ten per cent of children without type 1 diabetes-related autoantibodies at diagnosis in the Finnish Pediatric Diabetes Register. Diabetologia 66(3):438–449. https://doi.org/10.1007/s00125-022-05834-y
Estalella I, Rica I, Perez de Nanclares G et al (2007) Mutations in GCK and HNF-1ɑ explain the majority of cases with clinical diagnosis of MODY in Spain. Clin Endocrinol (Oxf) 67(4):538–546. https://doi.org/10.1111/j.1365-2265.2007.02921.x
Neu A, Feldhahn L, Ehehalt S, Hub R, Ranke MB (2009) Type 2 diabetes mellitus in children and adolescents is still a rare disease in Germany: a population-based assessment of the prevalence of type 2 diabetes and MODY in patients aged 0–20 years. Pediatr Diabetes 10(7):468–473. https://doi.org/10.1111/j.1399-5448.2009.00528.x
Małachowska B, Borowiec M, Antosik K et al (2018) Monogenic diabetes prevalence among Polish children—summary of 11 years-long nationwide genetic screening program. Pediatric Diabetes 19(1):53–58. https://doi.org/10.1111/pedi.12532
Delvecchio M, Mozzillo E, Salzano G et al (2017) Monogenic diabetes accounts for 63% of cases referred to 15 Italian pediatric diabetes centers during 2007–2012. J Clin Endocrinol Metab 102(6):1826–1834. https://doi.org/10.1210/jc.2016-2490
Carlsson A, Shepherd M, Ellard S et al (2020) Absence of islet autoantibodies and modestly raised glucose values at diabetes diagnosis should lead to testing for MODY: lessons from a 5-year Pediatric Swedish National Cohort Study. Diabetes Care 43(1):82–89. https://doi.org/10.2337/dc19-0747
Pihoker C, Gilliam LK, Ellard S et al (2013) Prevalence, characteristics and clinical diagnosis of maturity onset diabetes of the young due to mutations in HNF1A, HNF4A, and glucokinase: results from the SEARCH for Diabetes in Youth. J Clin Endocrinol Metab 98(10):4055–4062. https://doi.org/10.1210/jc.2013-1279
Shepherd M, Shields B, Hammersley S et al (2016) Systematic population screening, using biomarkers and genetic testing, identifies 2.5% of the U.K. Pediatric Diabetes Population With Monogenic Diabetes. Diabetes Care 39(11):1879–1888. https://doi.org/10.2337/dc16-0645
Shepherd M, Shields B, Ellard S, Rubio-Cabezas O, Hattersley AT (2009) A genetic diagnosis of HNF1A diabetes alters treatment and improves glycaemic control in the majority of insulin-treated patients. Diabet Med 26(4):437–441. https://doi.org/10.1111/j.1464-5491.2009.02690.x
Farahani P (2017) Non-severe hypoglycemia risk difference between sulfonylurea and sodium-glucose cotransporter-2 inhibitors (SGLT2-I) as an add-on to metformin in randomized controlled trials. J Popul Ther Clin Pharmacol 24:e32–e40. https://doi.org/10.22374/1710-6222.24.2.6
Bjørkhaug L, Bratland A, Njølstad PR, Molven A (2005) Functional dissection of the HNF-1alpha transcription factor: a study on nuclear localization and transcriptional activation. DNA Cell Biol 24(11):661–669. https://doi.org/10.1089/dna.2005.24.661
Funding
Open access funding provided by University of Bergen (incl Haukeland University Hospital)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Acknowledgements
We wish to thank the participants and their families for contributing to research on monogenic diabetes. We are indebted to biomedical laboratory scientist L. Aasmul for her help with clinical samples and testing, and staff engineer M. Ringdal for genetic analyses of DNA samples from probands and family members, both affiliated at the Mohn Center for Precision Diabetes Medicine. Some of the data were presented as an abstract at the International Society for Pediatric and Adolescent Diabetes (ISPAD) conference in Abu Dhabi, 13 October 2022.
Data availability
The datasets analysed during the current study are available from the corresponding author on reasonable request.
Funding
This work was supported by grants (to PRN) from the European Research Council (AdG no. 293574), the Bergen Research Foundation (‘Utilising the Mother and Child Cohort and the Medical Birth Registry for Better Health’), Stiftelsen Trond Mohn Foundation (Mohn Center of Diabetes Precision Medicine), the University of Bergen, Haukeland University Hospital, the Research Council of Norway (FRIPRO grant no. 240413), the Western Norway Regional Health Authority (Strategic Fund ‘Personalised Medicine for Children and Adults’), the Novo Nordisk Foundation (grant no. 54741) and the Norwegian Diabetes Association. This work was partly supported by grants (to PS) from the Norwegian Diabetes Association, Aarskog’s Foundation and the General Practitioner’s Foundation.
Authors’ relationships and activities
The authors declare that there are no relationships or activities that might bias, or be perceived to bias, their work.
Contribution statement
PS, AK, JM, JVS, PRN, ET, LB, AM and IA were involved in study design and development of study procedures. BBJ and SJ designed the NGS assay and performed the NGS analyses. JM and IA conducted the variant interpretation. PS, PRN, ET and TS were involved in sample collection, data collection, physiological investigations, clinical interpretation and patient care. TS, ES and LK were involved in participant enrolment. AK, MHS, JM, IA and LB designed, performed and interpreted the functional assays. EV contributed with design of figures (created by using elements and graphics from BioRender.com [licensed use]). All authors were involved in data interpretation and critically revising the report, and reviewed and approved the report for submission. PRN is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Svalastoga, P., Kaci, A., Molnes, J. et al. Characterisation of HNF1A variants in paediatric diabetes in Norway using functional and clinical investigations to unmask phenotype and monogenic diabetes. Diabetologia 66, 2226–2237 (2023). https://doi.org/10.1007/s00125-023-06012-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-023-06012-4